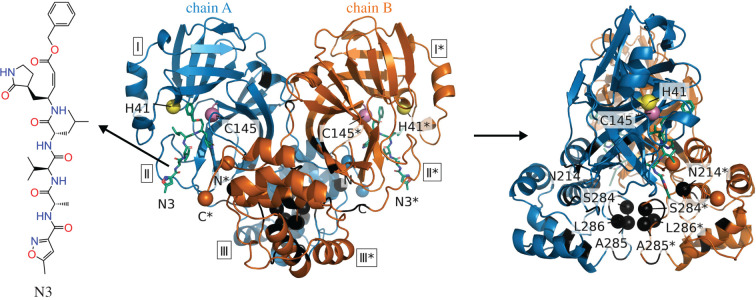
Figure 1.
The crystal structure of the SARS-CoV-2 Mpro with N3 inhibitor. Chain A of the dimer is shown in blue, while chain B, in orange. Domains are labelled with boxed Roman numerals (I, II and III). Amino acid residues of the catalytic dyad are indicated as yellow spheres for H41 and magenta spheres for C145. Asterisks mark residues from chain B (orange). Chain termini are shown as spheres and labelled N and C for chain A (blue) and N* and C* for chain B (orange). The N3 inhibitor is shown as green sticks. Experimentally identified dynamically allosteric residues N214 and SAL284-286 are shown as labelled black spheres. Identified control residue candidates for dynamic allostery in the SARS-CoV-2 Mpro that are distant from the catalytically active residues H41 and C145 are coloured in black on both homodimeric chains. These residues can be found in table 1.