Figure 1 |.
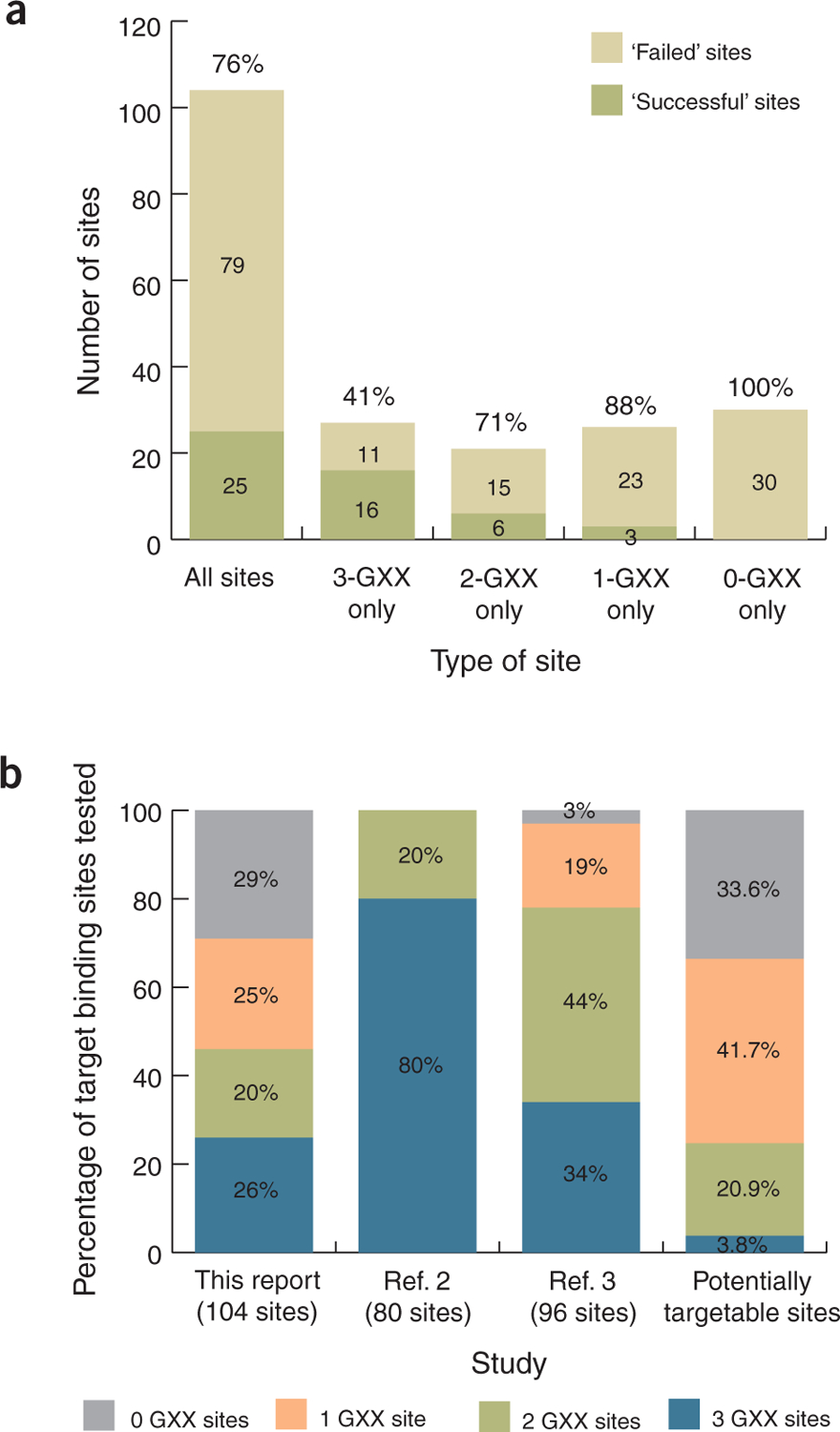
Large-scale evaluation of the modular assembly method for engineering zinc-finger arrays. (a) Data for success and failure of modular assembly as judged by the B2H assay for all 104 target DNA sites (see Supplementary Discussion for definition of success threshold) and for subsets of target sites containing three, two, one or no GXX subsites. ‘Successful’ sites are those for which at least one functional zinc-finger array was identified; ‘failed’ sites are those for which we failed to obtain a single successful array (see Supplementary Table 2 for details). Predicted failure rates for each set of target sites are indicated above the bars. (b) Distributions of target DNA sites used in different modular assembly evaluation studies according to the number of GXX subsites present. The distribution of all 107,011 potential 9-bp sites that can be targeted using 141 zinc-finger modules from the Zinc Finger Consortium Modular Assembly Kit 1.0 (ref. 6) is also shown (far right bar).
