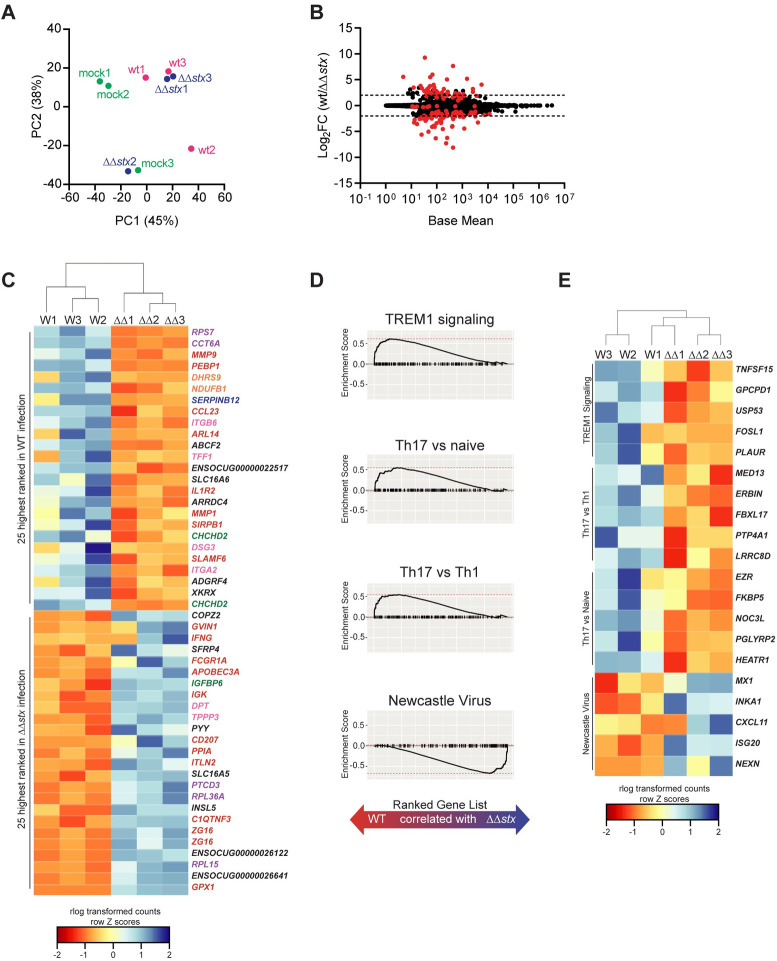
Fig 3. Profiles of colonic lamina propria cell transcriptional responses differ between animals infected with WT and ΔΔstx EHEC.
(A) Principal component analysis of rlog-transformed expression values of lamina propria cells. (B) Average expression level (base mean) and log2 fold change of transcript abundance from rabbits inoculated with WT or ΔΔstx EHEC. Red dots are genes with significantly different (p<0.05) transcript abundance. Dashed line indicates log2 fold change >2 or <-2. (C) Heat map of rlog-transformed read counts for top 25 and bottom 25 genes by rank. Rows are Z-normalized. Gene names are colored by function: red–immune, orange–metabolism, green–proliferation/apoptosis, blue–coagulation, purple–translation/protein folding, pink–barrier function/cytoskeleton, black–uncharacterized or other. (D) Gene set enrichment plot for selected pathways. Black tick marks are genes within pathway organized by rank. (E) Heat map of rlog-transformed read counts for top 5 genes in the leading edge for indicated pathway organized by rank.