Abstract
Chinese hamster ovary (CHO) cells are currently the primary host cell lines used in biotherapeutic manufacturing of monoclonal antibodies (mAbs) and other biopharmaceuticals. Cellular energy metabolism and endoplasmic reticulum (ER) stress are known to greatly impact cell growth, viability, and specific productivity of a biotherapeutic; but the molecular mechanisms are not fully understood. The authors previously employed multi-omics profiling to investigate the impact of a reduction in cysteine (Cys) feed concentration in a fed-batch process and found that disruption of the redox balance led to a substantial decline in cell viability and titer. Here, the multi-omics findings are expanded, and the impact redox imbalance has on ER stress, mitochondrial homeostasis, and lipid metabolism is explored. The reduced Cys feed activates the amino acid response (AAR), increases mitochondrial stress, and initiates gluconeogenesis. Multi-omics analysis reveals that together, ER stress and AAR signaling shift the cellular energy metabolism to rely primarily on anaplerotic reactions, consuming amino acids and producing lactate, to maintain energy generation. Furthermore, the pathways are demonstrated in which this shift in metabolism leads to a substantial decline in specific productivity and altered mAb glycosylation. Through this work, meaningful bioprocess markers and targets for genetic engineering are identified.
Keywords: Chinese hamster ovary bioprocessing, energy metabolism, endoplasmic reticulum stress, monoclonal antibodies, multi-omics, redox
1. Introduction
Monoclonal antibodies (mAb) are a major class of biotherapeutics that have demonstrated great effectiveness in the treatment of a variety of diseases.[1] Chinese hamster ovary (CHO) cells have become the primary host for biopharmaceutical therapeutic production due to their ability to properly fold proteins and incorporate the necessary post-translational modifications.[2,3] Today, mAbs are manufactured in fed-batch and continuous bioprocesses in which amino acids and other nutrients are fed to maintain growth and productivity.[4]
Multi-omics analysis is rapidly becoming an important strategy for gaining a deeper understanding of the cellular biology during bioprocessing and is increasingly being used in CHO bioprocess development.[5] The combination of transcriptomics, proteomics, and metabolomics has been shown to enhance bioprocess understanding and improve titers.[6,7] More recently, lipidomics, an emerging ‘omics field, has been shown to enhance the understanding of CHO cellular bioprocesses, especially in pathways related to energy metabolism and storage.[8]
Cysteine (Cys) is an amino acid that plays a crucial role in the production of antioxidant molecules such as glutathione (GSH), which subsequently impacts protein folding pathways and other important cellular processes. Therefore, limitation in Cys can potentially have far reaching effects. In some mammalian cells, Cys can be produced from the precursor amino acids—serine and methionine.[9] In a previous study, we performed multi-omics analysis (transcriptomics, proteomics, and metabolomics) over the time course of a CHO bioprocess to explore the biological changes that led to a dramatic reduction in viability and mAb titer when the Cys feed concentration was reduced by 15%, compared to a Cys control concentration, in a fed-batch bioprocess.[10] Pathway analysis indicated that low expression of the Cys biosynthesis enzymes (CBS and CTH) limited the ability of the cells themselves to produce Cys, and thus, the bolus feed delivered daily was required to maintain the intracellular Cys level. As a result, the intracellular concentration of Cys between individual feed additions in the low Cys condition was found to be depleted to quite low levels prior to the daily feed. Interestingly, Cys is generally considered to be a semi-essential amino acid.[11] The depletion of intracellular Cys during mAb production was found to have multiple deleterious effects, including severe reduction in antioxidant molecules resulting in oxidative stress. The resultant redox imbalance led to enhanced endoplasmic reticulum (ER) stress, apoptosis, and reduced mAb titer.[10]
In this paper, using multi-omics (including lipidomics) analysis of the same samples collected in the previous study,[10] we delve more deeply into the impact of the 15% reduction of Cys feed, relative to the control, focusing on the negative effects on cellular energy metabolism and resultant specific productivity. Highly productive cell lines will always generate some ER stress; however, additional and thus excess stress will impact cell viability, productivity, and product quality.[12] Moreover, high bioprocess productivity is known to be significantly linked to oxidative phosphorylation, that is, energy generation through the tricarboxylic acid (TCA) cycle and electron transport chain (ETC) in the mitochondria.[13] At the same time, reduction or depletion of essential (or semi-essential) amino acids within a cell is known to cause stress and trigger unfavorable metabolic changes, that is, the amino acid response (AAR).[14,15] The challenge is to maintain the CHO cells in a highly energetic metabolic state to maximize protein production while at the same time not generate excessive stress that reduces productivity.
Using multi-omics for examination of known metabolic and lipidomic pathways, we map out key events resulting from the limitation in Cys within the cell leading to enhanced ER stress, oxidative stress, decreased TCA cycle activity (mitochondrial dysfunction), autophagy, and ultimately cell death. The results of this paper lead to an increased understanding of the cell biology involved in the production of a mAb. Through this understanding, we uncover potential bioprocess biomarkers and new targets for genetic engineering, all potentially leading to improved bioprocess productivity.
2. Results
2.1. Low Cysteine Feed Concentration Leads to a Decrease in Titer, Specific Productivity, and Product Quality
5 L bioreactors were used to cultivate DG44 CHO cells for 14 days in a fed-batch bioprocess with daily bolus feeding of nutrients, as described previously.[10] Three separate Cys feed concentrations were utilized in triplicate for a total of nine bioreactor runs. Note that the same collected samples from our previous study were used for the multi-omics analysis in the present paper. In our previous paper,[10] we found that only a 15% reduction in the Cys feed concentration (relative to control) led to a halving of the mAb titer (see Figure 1A). Sampling was conducted just prior to the daily bolus addition (indicated by black arrows in Figure 1A); therefore, the measured abundances for the various ‘omics methods represent the lowest intracellular concentrations experienced before the next feeding. Previously, we found depletion of cellular antioxidants, especially GSH, in the low Cys condition, which in turn caused increased oxidative stress and, ultimately, cell death.[10] As the GSH was consumed over the 24 h between the daily bolus feeds, the antioxidants were not depleted, on average, as much for the control and high Cys media conditions compared to the low Cys condition.
Figure 1.
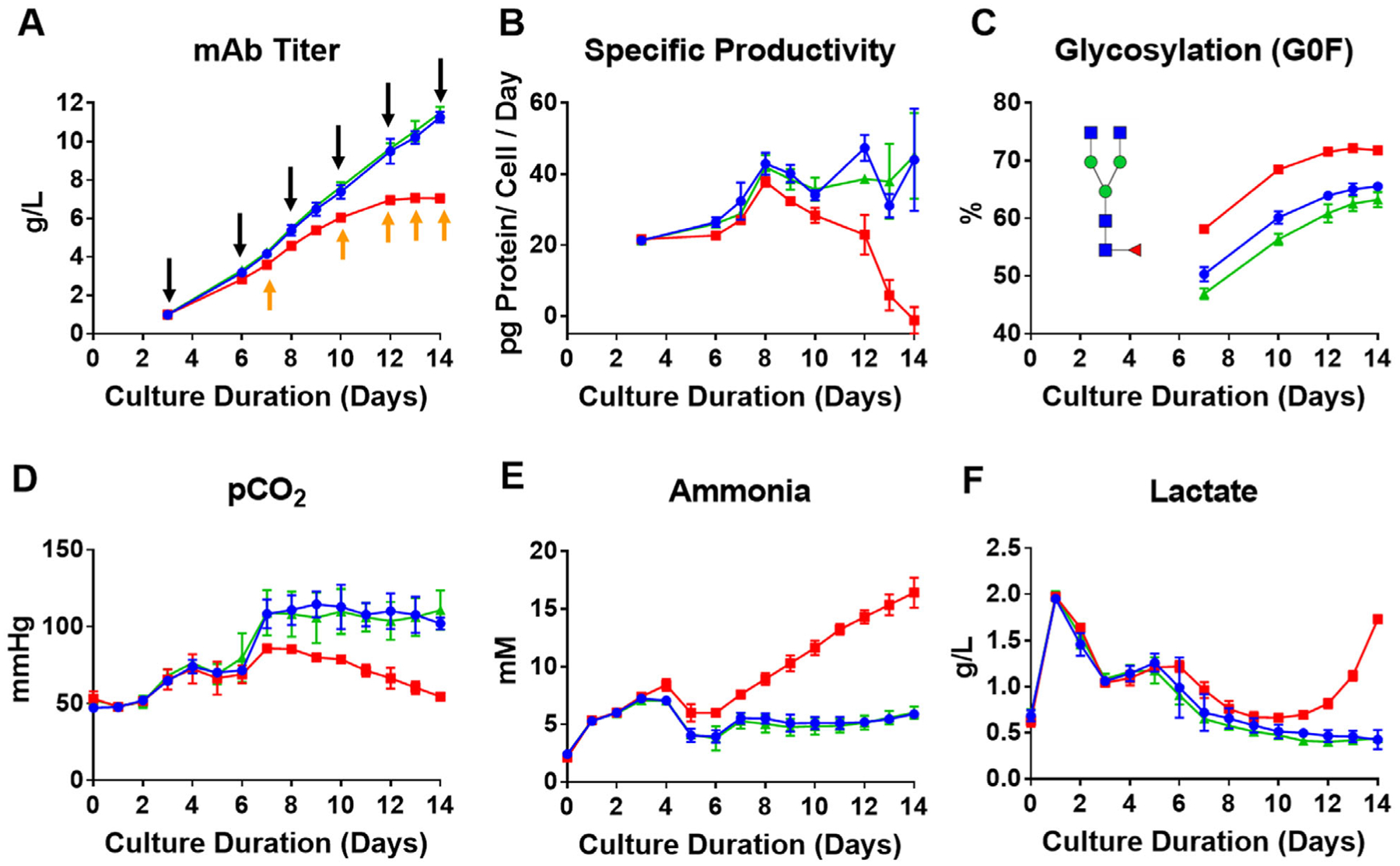
Cell culture bioprocess parameters demonstrating changes in metabolism and productivity when insufficient levels of Cys are added to the feed. Red, blue, and green represent, respectively, the three Cys feed concentrations low (−15%), control, and high Cys (+20%). A) Therapeutic mAb titer (black arrows indicating when ‘omics samples were taken and orange arrows for glycosylation analysis prior to the next daily feeding); B) specific productivity plot as a function of culture time; C) relative glycosylation distribution of the therapeutic mAb as a function of culture time showing a relative increase in the G0F glycoform in the low Cys condition; D) partial pressure of CO2 as a function of culture time; E) concentration of extracellular ammonia as a function of culture time; and F) concentration of extracellular lactate as a function of culture time. Data plotted represent the average of three separate bioprocess runs with error bars indicating the standard deviation.
As seen in Figure 1B, specific productivity was found to decrease significantly beginning on day 8 in the low Cys condition, ultimately reaching zero specific productivity at the end of the cultivation on day 14. Additionally, product quality of the mAb was affected; specifically, the N-linked glycosylation profile changed where the G0F glycoform, on average, increased for the low Cys feed (Figure 1C).
Relative to the control and high Cys feed conditions, changes in widely used metabolic markers were also observed in the low Cys condition, for example, decrease in the partial pressure of CO2 (pCO2) (50% by day 14, Figure 1D), as well as elevated levels of ammonia (NH4 +) (threefold higher by day 14, Figure 1E) and lactate (fourfold higher by day 14, Figure 1F), all measured extracellularly. The decrease in pCO2 and increase in NH4 + suggest that cells that are fed only 15% less Cys than the control exhibited a significant unfavorable shift in energy metabolism.
Ammonia concentration is known to affect mAb N-glycosylation.[16] We observed no change in RNA expression or protein abundance of the enzyme beta-1,4-galactosyltransferase (B4GALT), which is responsible for attaching the galactose sugar from the UDP-galactose precursor (Figure S1, Supporting Information). Therefore, the change in intracellular pH due to the increased level of ammonia is likely the main driver behind the decreased activity of B4GALT, leading to increased levels of the G0F glycoform of the mAb (Figure 1C), in agreement with the results of others.[17]
We focus below on the unfavorable shift in energy metabolism for the low Cys feed. Our results reveal a collection of multifaceted and interconnected cellular stress responses such as redox imbalance, ER stress, amino acid limitation, and mitochondrial dysfunction when the Cys level was reduced.
2.2. Endoplasmic Reticulum Stress
Typically, CHO cells used in bioprocessing are genetically engineered to maximize mAb or other biopharmaceutical expression; this high productivity inevitably leads to some ER stress, and the unfolded protein response (UPR) is generally activated by virtue of enhanced protein synthesis and processing in the ER.[18] In this study, in the low Cys fed condition, compared to the control and high Cys feeds, we observed as much as a twofold increase in RNA expression of the endoribonuclease, inositol requiring enzyme-1a (IRE1a), a known marker of ER stress (Figure 2A).[19] This result is in agreement with our previous findings where increased expression of the chaperon protein BiP was observed when the Cys level was limited, indicative of enhanced UPR.[10]
Figure 2.
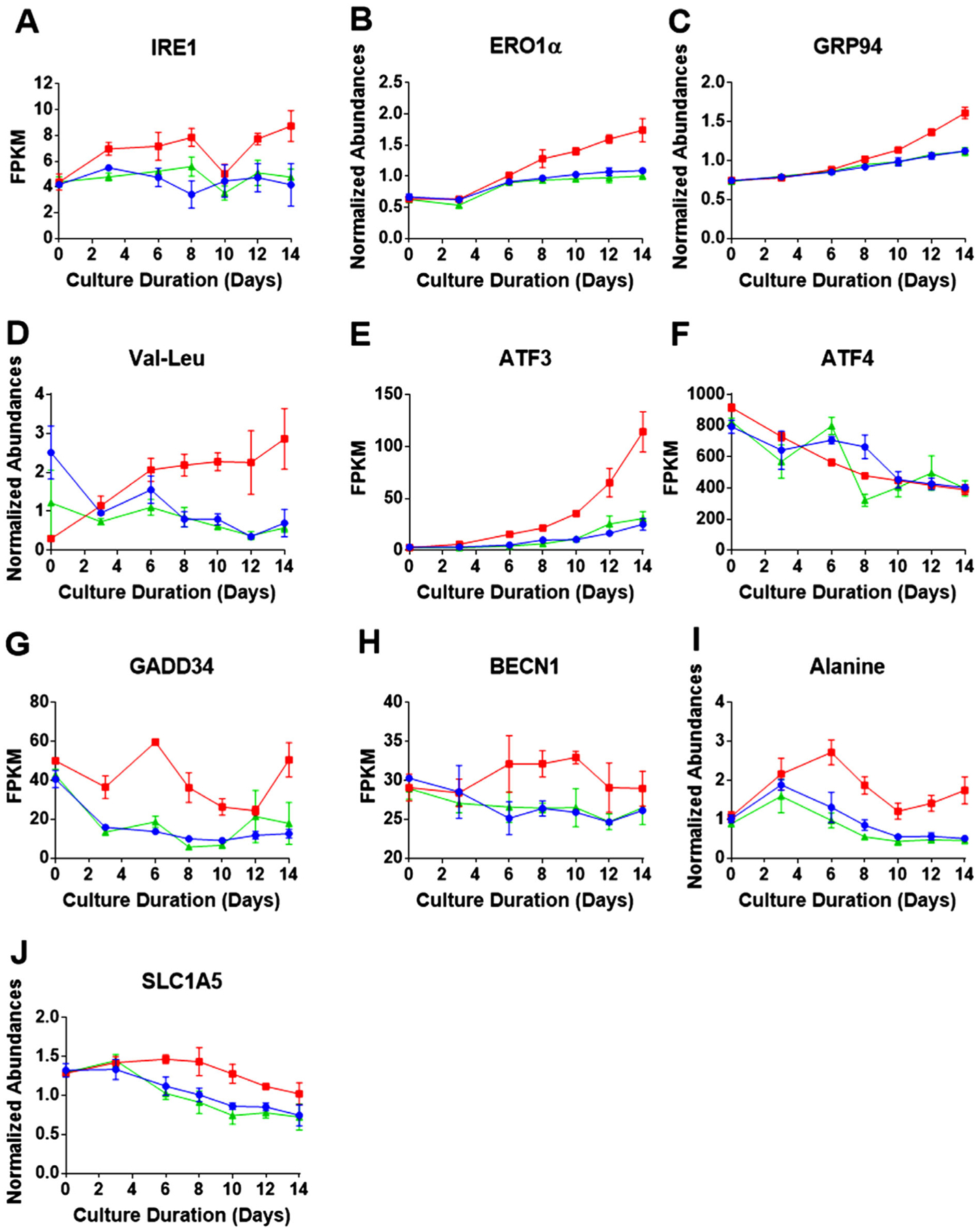
Intracellular time course profiles for proteins and RNA transcripts related to ER stress, UPR, and autophagy. Red, blue, and green represent the three Cys feed concentrations low, control, and high Cys, respectively. Over the time course of the bioprocess, A) shows RNA profile of ER stress sensor IRE1; B) protein abundance of ERO1α (a member of the oxidative protein folding machinery); C) protein abundance of chaperon protein GRP94; D) abundance of Val-Leu dipeptide; E) RNA expression of ATF3 (response factor activated by the UPR); F) RNA expression of ATF4 (response factor activated by the UPR); G) RNA expression of GADD34 (an indicator of growth arrest); H) RNA expression of BECN1 (an indicator of apoptosis); I) relative abundance of the amino acid alanine; and J) glutamine transporter SLC1A5 protein abundance (an indicator of glutaminolysis). Data shown represent the average of three bioprocesses with the error bars indicating the standard deviation (RNA transcripts (FPKM) and proteins (abundances)) for each of the Cys feed conditions.
Since we previously found depletion of GSH, a major antioxidant, under the low Cys condition,[10] significant disruption of redox homeostasis can be expected. In particular, decreased levels of GSH can lead to redox imbalance in the ER, affecting the enzymes that control disulfide bond formation, resulting in increased protein misfolding.[20] Cells are known to respond to the increased protein misfolding in the ER by upregulating the oxidative protein folding pathway, which involves two proteins: protein disulfide-isomerase (PDI) that rearranges incorrect disulfide bonds and ER oxidoreductin 1α (ERO1α) that reoxidizes PDI, while generating H2O2 in the process.[21] Interestingly, as seen in Figure 2B, the level of ERO1α abundance was found to be higher in the low Cys condition, relative to the control and high, as early as day 6, indicating that the cells are reacting to the enhanced ER stress caused by the lack of GSH. We also observed increased protein abundance of chaperon proteins, in addition to the previously cited BiP,[10] for example, heat shock protein 90 kDa (GRP94) (see Figure 2C), a further indication of the increased UPR response in the low Cys condition.
The inability to fold proteins properly in the ER will lead to misfolded mAb proteins that need to be degraded to prevent damage to the cells and to recycle the amino acids. In the low Cys condition, relative to the control, we observed evidence of enhanced protein degradation in the form of increased levels of dipeptides. As examples, the time course profile of the dipeptide, valine-leucine is shown in Figure 2D, while other dipeptides can be found in Figure S2, Supporting Information.
2.3. Cellular Stress: Amino Acid Response and ER Stress
As we have shown in our previous paper,[10] when insufficient levels of Cys were fed, the intracellular concentration of this semi-essential amino acid decreased to very low levels until the addition of the next bolus feed. Additionally, amino acid analysis of the supernatant at the end of the feed period showed depletion of Cys in the media (data not shown). Due to the intracellular depletion of Cys, the amino acid response (AAR) was activated, signaling a sense of starvation in the cell. The expressions of the activating transcription factors 3 and 4 (ATF3 and ATF4) are known to be upregulated due to the activation of both the AAR and ER stress.[14] In our experiments, we found steadily increasing levels of RNA expression over the bioprocess cycle for ATF3 in the low Cys condition, relative to the control and high levels (Figure 2E). Interestingly, RNA levels of ATF4 remained similar for all three Cys feed conditions (Figure 2F). The significant increase in ATF3 in Figure 2E points to the increased stress in the low Cys fed CHO cells, relative to the control and high Cys feed, and ultimately to increased apoptosis.[22]
Activation of the AAR and increased ER stress initiates a variety of cellular responses, including gluconeogenesis, growth arrest, autophagy, and apoptosis, as well as decreasing global protein synthesis.[14,15] All of these responses are detrimental to the specific productivity of the bioprocess (see Figure 1B). In the low Cys fed condition, we observed an increase in the DNA damage-inducible protein (GADD34) (Figure 2G) and expression of the Beclin1 gene (BECN1) (Figure 2H). These trends suggest cellular growth arrest and an increase in autophagy.[23] We next turn to an examination of the effect of ER stress and AAR on the energy metabolism of the cell.
2.4. Impact of ER Stress and AAR on Mitochondrial Function
The TCA cycle within the mitochondria is a cornerstone of cellular energy metabolism as it not only produces precursors to enable the ETC to generate ATP, but the cycle also provides the cell with the flexibility to adapt to environmental changes.[24] As shown in the schematic overview of the energy metabolism pathways in Figure 3, cells typically convert pyruvate, generated from glycolysis, to acetyl-CoA, the latter being the entry point into the mitochondria and the TCA cycle. Increased ER stress and activation of the AAR generates mitochondrial stress, affecting the TCA cycle and thus, the global energy metabolism in the cell. Figure 3i,iv highlight the anaplerotic reactions in which amino acids and protein catabolism replenishes TCA cycle intermediates. A high-level overview of glycolysis is shown in Figure 3ii, while Figure 3iii presents the overall connection between the TCA cycle and lipid metabolism.
Figure 3.
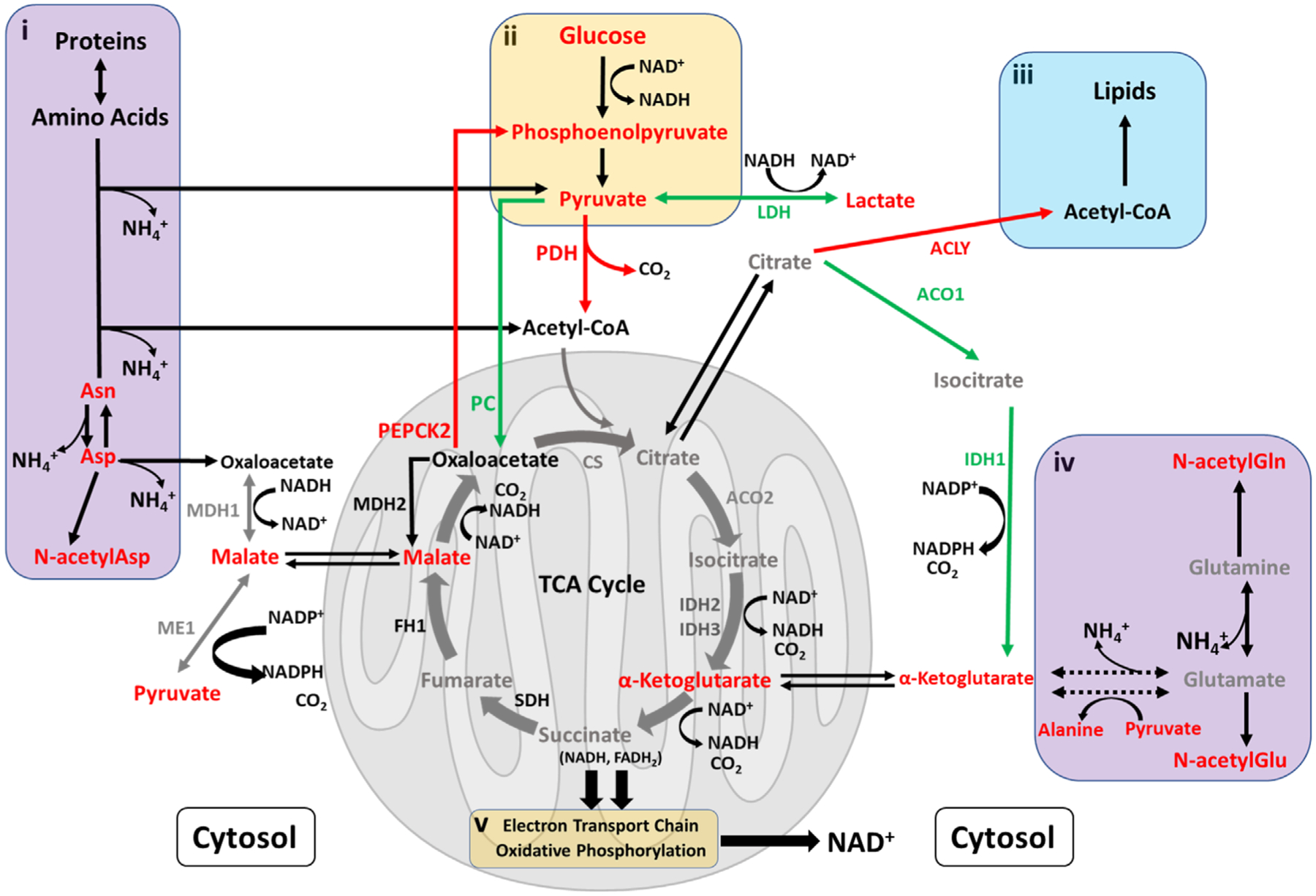
Schematic overview of pathways related to energy metabolism. Red represents an increase (>1.2-fold) in the ratio of the low to control Cys feed condition on day 14, while green indicates a decrease (<0.8-fold). Gray represents no change while black indicates data were not available. In the figure, metabolic enzymes (abbreviated names) are included. Separate insets illustrate: i) breakdown of amino acids and proteins to replenish TCA cycle intermediates, with the release of ammonia as a by-product; ii) abbreviated overview of glycolysis; iii) interconnectivity between lipid metabolism and the TCA cycle; and iv) the well-known glutaminolysis process to support the TCA cycle, with release of ammonia.
Oxaloacetate is an important TCA cycle intermediate whose level can be controlled by two enzymes—pyruvate carboxylase (PC) and phosphoenolpyruvate carboxykinase 2 (PEPCK2).[15] As seen in Figure 3, PC catalyzes the production of oxaloacetate from pyruvate while PEPCK2 removes oxaloacetate from the TCA cycle with conversion to phosphoenolpyruvate. The increase in transcription factor ATF3 (Figure 2E) is known to increase expression of the PCK2 gene encoding for the enzyme PEPCK2.[15] In the low Cys condition, the RNA expression of the PCK2 gene was found to be higher, as early as day 3, compared to the control and high Cys conditions (Figure 4A), leading to increased enzyme abundance of PEPCK2 for the low Cys feed (Figure 4B). This upregulation in protein abundance can increase the removal of oxaloacetate, leading to a reduction in the TCA cycle activity. At the same time, for the low Cys feed, we observed a decrease in RNA expression of the PC enzyme (Figure 4C), resulting in a decrease in PC enzyme abundance (Figure 4D) and likely a reduction in oxaloacetate. As seen in Figure 3, with limited oxaloacetate, the enzyme citrate synthetase (CS) cannot sufficiently use acetyl-CoA to maintain the TCA cycle; this results in lower levels of NADH and FADH2, molecules that are required for ATP generation by the ETC.[25]
Figure 4.
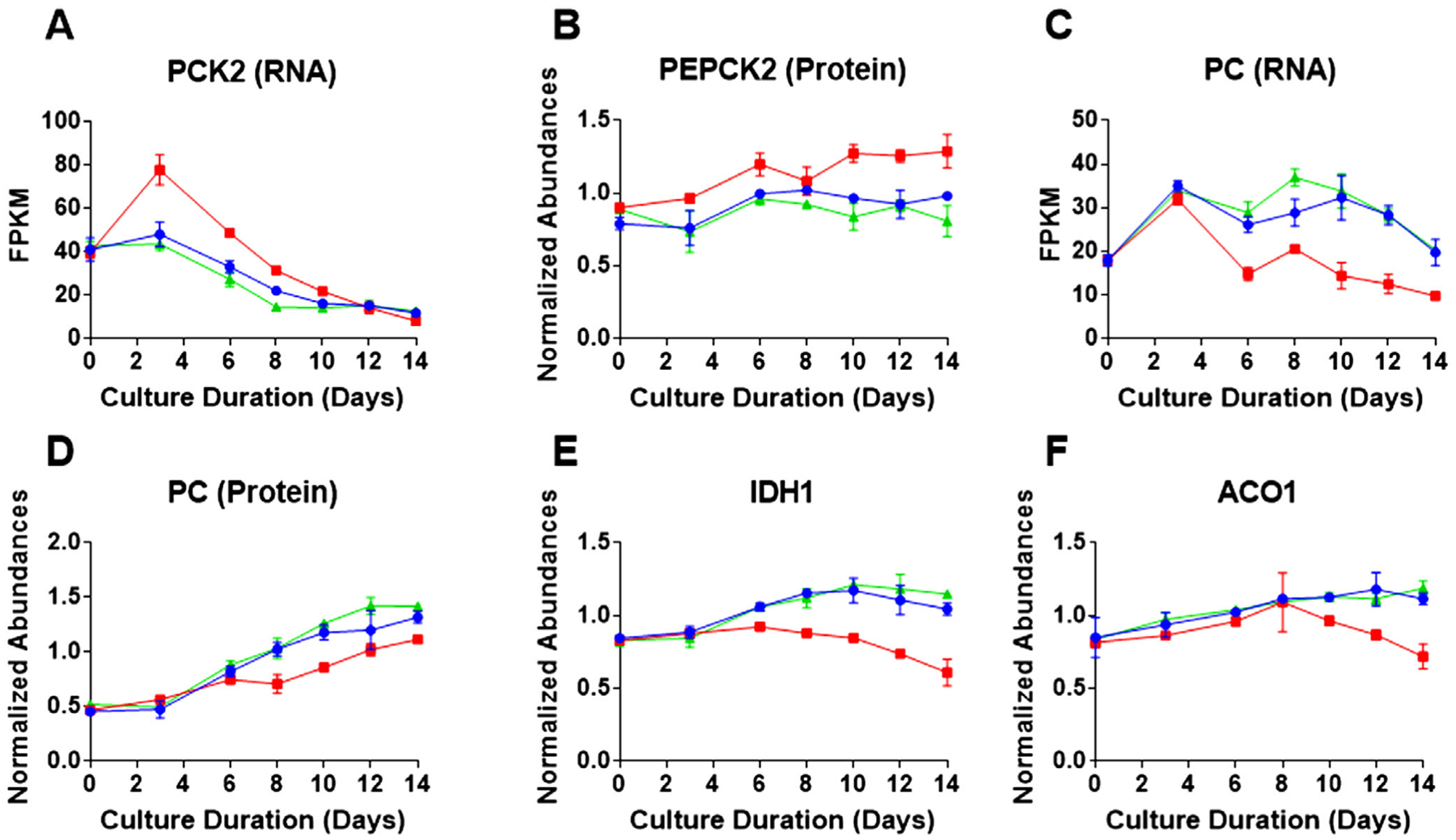
Intracellular time course profiles for proteins and metabolites related to the TCA cycle and energy metabolism. Red, blue, and green represent the three Cys feed concentrations low, control, and high Cys, respectively. Data shown represents the average protein or metabolite abundance for three bioprocess runs with the error bars indicating standard deviation for each of the Cys feed conditions as a function of culture duration for A) PCK2 RNA transcript level; B) PEPCK2 relative protein abundance profile; C) pyruvate carboxylase (PC) RNA level; D) PC relative protein abundance; E) NADPH-dependent isocitrate dehydrogenase (IDH1) protein abundance; and F) NADPH-dependent aconitase (ACO1) protein abundance.
With reduction in the TCA cycle, the cell tries to generate the necessary cellular energy by shifting to an alternative anaplerotic pathway to maintain the cycle. The breakdown of glutamine and glutamate to produce α-ketoglutarate, an intermediate in the TCA cycle, either through the release of ammonia or through the transaminase reaction producing alanine,[26] are well-known anaplerotic reactions (Figure 3). Evidence for this anaplerotic pathway is seen in the increase in ammonia as the cultivation proceeds (Figure 1E) and intracellular alanine (Ala) (Figure 2I). Furthermore, we detected increased expression of the glutamine transporter SLC1A5 (Figure 2J), a known glutaminolysis indicator.[27]
Cells can also generate α-ketoglutarate through a separate pathway in the cytoplasm using citrate by means of IDH1 and ACO1 (NADP+ dependent), as shown in Figure 3. Figure 4E,F present the time course protein abundance profiles for IDH1 and ACO1, respectively. Both enzymes are found to decrease dramatically over time under the low Cys condition, relative to the control and high Cys feed. It appears that the cells fed insufficient levels of Cys were less able to generate α-ketoglutarate from citrate and relied heavily on glutaminolysis to replenish the TCA cycle intermediates.
Returning to the removal of oxaloacetate from the TCA cycle, higher levels of phosphoenolpyruvate were found in the low Cys condition, compared to the control and high, signaling gluconeogenesis, as shown in Figure 5B. Additionally, sugars in the polyol pathway, for example, fructose and sorbitol were found to be elevated in the low Cys condition. The time course plot shows significant increases of fructose after day10 (Figure 5D); other related sugars are presented in Figure S4, Supporting Information. Ribose was also found to be higher in the low Cys fed condition resulting from increased gluconeogenesis (Figure 5E). Coupled with the continuous addition of glucose to the bioreactor as a part of the nutrient feeding, gluconeogenesis contributed to elevated levels of pyruvate (along with AAR resulting in lower expression levels of the PC enzyme) for the low Cys condition (Figure 5C). Also seen are increases in other glycolysis intermediates such as 3-phosphoglycerate (3PG) and 2-phosphoglycerate (2PG) (Figure 5A and Figure S3, Supporting Information).
Figure 5.
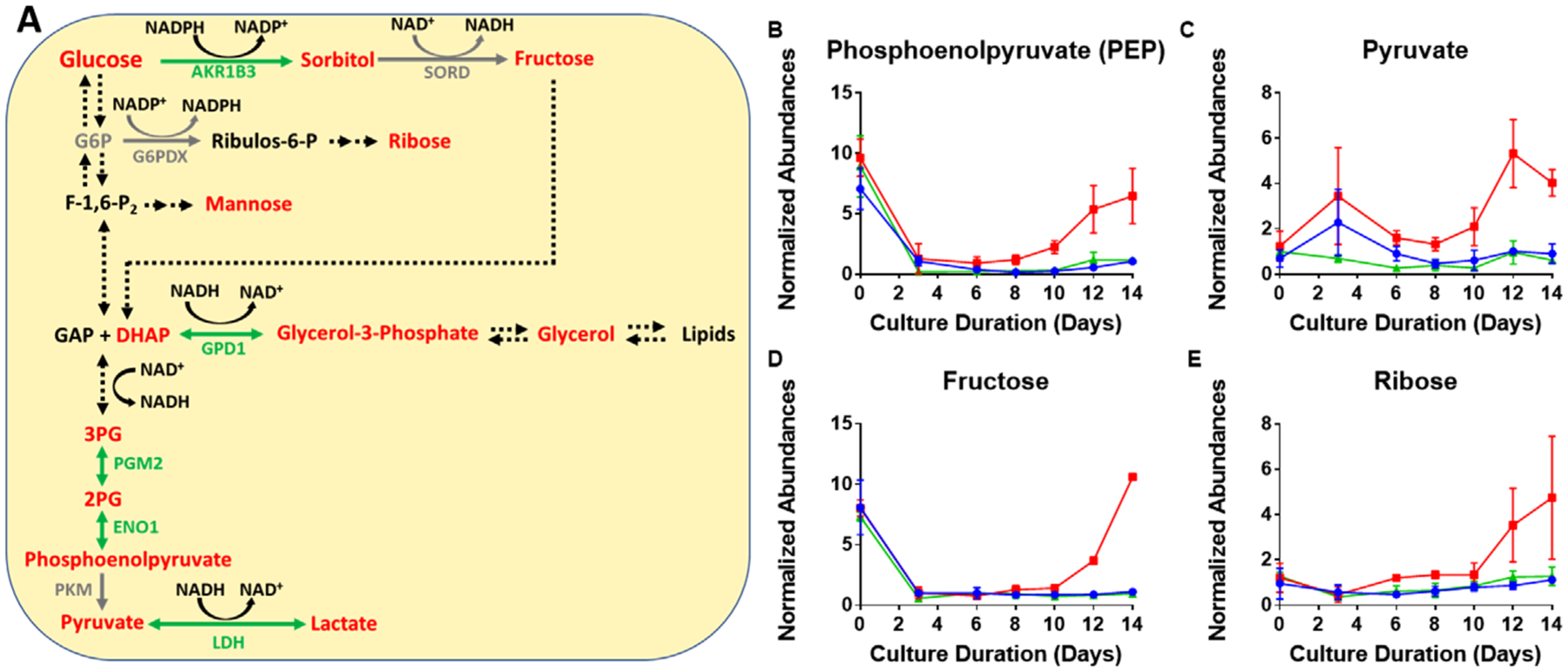
Impact of insufficient Cys feeding on glycolysis. A) A detailed picture of the glycolysis pathway shown in Figure 2. Red represents an increase (>1.2-fold) in the ratio of the low to control Cys feed condition on day 14, while green indicates a decrease (<0.8-fold). Gray represents no change while black indicates data were not available. Abbreviated protein names are shown. Average time course profiles for intracellular metabolites related to glycolysis are shown; B) phosphoenolpyruvate (PEP) abundance; C) pyruvate abundance; D) fructose abundance; and E) ribose abundance. The error bars indicate standard deviation of abundance for three bioprocess runs. Red, blue, and green lines represent the three Cys feed concentrations low, control, and high Cys, respectively.
2.5. Lipid Metabolism
Redox imbalance and Cys limitation, as we have pointed out, can enhance oxidative stress and induce autophagy. Due to the importance of lipids in membrane composition as well as energy storage and generation, we investigated changes in the lipidome during bioprocessing to explore the impact of insufficient Cys feeding and resultant redox imbalance on lipid metabolism. Figure 6A presents an overview of the lipid metabolism pathway,[28] demonstrating the breakdown and biosynthesis of glycerophospholipids and glycerolipids. Figure 6B illustrates an example of how a specific phosphatidylethanolamine (PE) with two 18 carbon fatty acid chains and one double bond is degraded. Compared to the control and high Cys conditions, we observed increased glycerophospholipid membrane degradation to lysophospholipids in the low Cys condition. For example, we found higher levels of plasmalogen PE (P-18:1) and lyso-PE (18:1), both of which are breakdown products of glycerophospholipid degradation (Figure 6C,D). As oxidative stress and autophagy persists, the second fatty acid chain can be cleaved from the lysophosphatidylethanolamine and lysophosphatidylcholine leaving only the glycerophosphate groups glycerophosphoethanolamine (GPE) and glycerophosphocholine (GPC). Figure 6E,F show that the intracellular levels of GPE and GPC in the low Cys condition increased dramatically over the bioprocess production time, reaching a level almost 80 times higher than the control condition. This substantial increase in GPE and GPC inside the cell is due to the fact that these products are common for all fatty acid chain lengths and levels of unsaturation.
Figure 6.
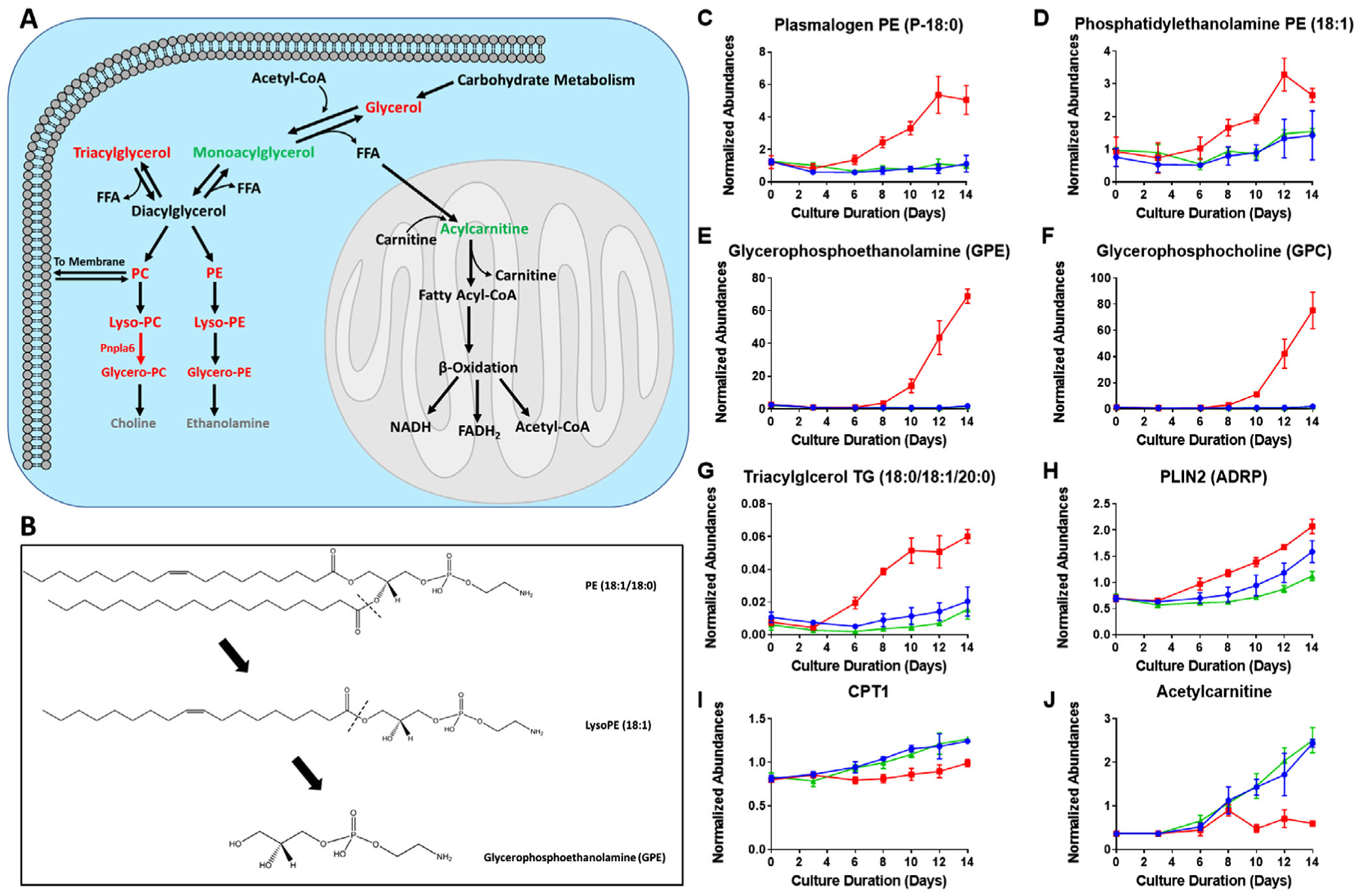
Impact of insufficient Cys feeding on glycolysis and related intracellular metabolites. A) A detailed picture of the lipid metabolism pathway shown in Figure 2. Red represents an increase (>1.2-fold) in the ratio of the low to control Cys feed condition on day 14, while green indicates a decrease (<0.8-fold). Gray represents no change while black indicates data were not available. B) Mechanism of phospholipid degradation is shown. In C–J), the colors red, blue, and green represent the three Cys feed concentrations low, control, and high Cys, respectively. Average time course for intracellular lipids and related proteins are shown with error bars indicating standard deviation of relative abundance for three bioprocess runs: C) increase in plasmalogen PE (P-18:0) with a single fatty acid chain resulting from lipid degradation; D) lysophosphatidylethanolamine (lyso-PE (18:1)); E,F) glycerophospholipid headgroup GPE and GPC, respectively, released due to lipid degradation; G) accumulation of triacylglycerols; H) protein abundance profile of PLIN2, an indicator of lipid droplet (LD) biosynthesis; I) protein abundance profile of CPT1, enzyme responsible for acylcarnitine production; and J) acetylcarnitine which helps transport free fatty acids through the inner mitochondrial membrane for fatty acid β-oxidation.
As cells experience autophagy and increased membrane degradation due to increased oxidative and ER stress along with activation of the AAR, there will be an accumulation of free fatty acids within the cell. Increased levels of these free fatty acids can cause lipotoxicity[29]; therefore, cells will sequester excess fatty acids in neutral triacylglycerols (TG).[30] In our experiments, we observed accumulating levels of TGs over time in the low Cys condition, demonstrating that such cells are actively sequestering a large portion of the released free fatty acids. Figure 6G shows the accumulation over time in the low Cys condition of a specific TG (18:0/18:1/20:0); other triacylglycerol examples can be found in Figure S5, Supporting Information.
TG molecules are stored in lipid droplets (LDs) that are bound by a phospholipid membrane monolayer.[31] The enhancement in TG level is expected to increase the size and number of LDs in the cell.[28] Our results show evidence of LD biogenesis in the form of increased levels of the protein perilipin 2 (PLIN2) in the low Cys condition, a protein known to enhance lipid droplet formation (Figure 6H).[32] Since we observed that the TCA cycle was not generating sufficient energy in the low Cys condition, we expected β-oxidation of fatty acids to occur to produce ATP and supply cellular energy. However, surprisingly, we found that the cells stored the fatty acids in the form of TGs.
Acylcarnitine is a carrier that facilitates the transport of fatty acids through the inner mitochondrial membrane where β-oxidation would take place, as illustrated in Figure 6A.[33] We found a lower abundance of the important regulator of mitochondrial β -oxidation, carnitine palmitoyl transferase 1 (CPT1), the enzyme that catalyzes the production of acylcarnitine,[34] in the low Cys condition, compared to control and high Cys feed (Figure 6I). The reduction in acetylcarnitine during days 7 to 14 of the cultivation (Figure 6J) can be seen to be a result of the reduced abundance of the CPT1 enzyme. Thus, the transport of free fatty acids through the inner mitochondrial membrane was hindered.[35,36] Additionally, we found reduced protein abundance of long-chain acyl-CoA synthetase (LACS) (Figure S6A, Supporting Information), a protein that activates free fatty acids with the addition of coenzyme A for CPT1 transport into the mitochondria. LACS is regulated by the transcription factor peroxisome proliferator activated receptor PPARγ,[37] which was also found to be decreased in the low Cys condition (Figure S6B, Supporting Information).
3. Discussion
As shown in Figure 1, a small 15% reduction in Cys batch feed, relative to the control and 20% higher Cys level, resulted in the reduction of specific productivity to zero and halving of the titer at the end of the bioprocess on day 14. In our previous study,[10] we found that the reduced Cys feed led to depletion of cellular Cys and subsequently, GSH, a major antioxidant in the cell, as well as other antioxidants, for example, taurine and hypotaurine.[10] In the present work, using the same samples, we were able to explore in-depth by means of multi-omics, the effect of antioxidant depletion and redox imbalance, as well as the cellular reduction of Cys, a critical semi-essential amino acid, on the CHO cell biology.
During oxidative protein folding in the ER, GSH is known to play a key role in the reduction of the thiols on PDI, an enzyme that facilitates disulfide bond formation.[38] Although cells in the low Cys condition, relative to the control and high Cys feed levels, upregulated the oxidative protein folding pathway resulting from the redox imbalance, it is likely that the cells were unable to restore the misfolded/unfolded proteins to their correct structure for secretion, leading to ER stress over and above that already present due to the high productivity of the CHO cells.[39,40] Furthermore, as cells in the low Cys condition upregulated the re-oxidizing enzyme ERO1α (Figure 2B), enhanced formation of ROS in the ER could result, further increasing the ER stress as well as general cellular oxidative stress.[18]
The decline in bioprocess productivity observed in the low Cys condition has been found to be due to multiple factors. Cells in the low Cys condition, again relative to the other two conditions, increased protein degradation pathways to clear the accumulated misfolded proteins, as evidenced by the increase in dipeptides (Figure 2D and Figure S1, Supporting Information). Formation and degradation of misfolded proteins is one of the factors contributing to reduced specific productivity and product titer.
Enhanced ROS production coupled with the decrease in cellular antioxidants and severe ER stress will negatively impact the TCA cycle and ETC, thus causing mitochondrial dysfunction.[41–43] Consequently, as cells are fed an insufficient level of Cys, ATP generation through oxidative phosphorylation will be reduced. Furthermore, the equilibrium of NAD+/NADH will not be maintained by the ETC; thus, the cells will ultimately resort to lactate production (as seen after day 10, Figure 1F) in an attempt to improve the NAD+ levels. Increased lactate in the extracellular region, however, requires the addition of base to maintain the culture pH, leading to increased osmolality which is known to have a significant negative impact on cellular growth and mAb titer.[44]
Beyond ER and mitochondrial stress, insufficient Cys feeding and resultant intracellular Cys depletion led to the activation of the amino acid response (AAR) pathway. It is known that together, ER stress, mitochondrial stress, and amino acid limitation can lead to the cellular integrated stress response (ISR) where increased expression of the transcription factors ATF3 and/or ATF4 can result in reduced protein synthesis per cell, lower cell growth, initiation of gluconeogenesis, and apoptosis.[19] Although we did not observe an increase in ATF4 between the different Cys conditions, we did find a significant increase in ATF3 in the low Cys condition, as early as day 6, relative to the control and high Cys conditions (Figure 2). Others have demonstrated that an increase in ATF3 following the deprivation of an essential amino acid induced the ISR.[45] Although limitation in other amino acids (e.g., asparagine and serine) was found to impact cell growth in some CHO cells, the results varied based on cell line.[46] Interestingly, limitation in Cys consistently resulted in lower cell culture performance and the sensitivity to Cys did not appear to be cell line specific.[47]
In the low Cys condition, in comparison to the control and high level, the AAR impacted the TCA cycle functionality as the relative increased expression and protein abundance of the metabolic enzyme PEPCK2 converted oxaloacetate, an important TCA cycle intermediate, back to the glycolysis intermediate phosphoenolpyruvate. As shown in Figure 5, we observed an increased abundance of phosphoenolpyruvate and pyruvate as well as gluconeogenesis, as evidenced by the rising levels of sugars such as fructose starting on day 10, in the low Cys condition compared to the control and high Cys conditions. Even though the cells are constantly fed glucose, the cells in the low Cys condition still sensed some level of “starvation” as a result of the limited amount of Cys.
Due to the impact on the TCA cycle from the AAR and the ER stress in the low Cys condition, TCA cycle intermediates were replenished primarily through glutaminolysis, releasing ammonia as a by-product, as seen in Figure 1E. Interestingly, this behavior of increased gluconeogenesis and anaplerotic metabolism is analogous to rapidly multiplying tumor cells where such cells exhibit high rates of metabolism and compete with other cells for the limited available nutrients.[48,49] The reduction in pCO2 observed in the low Cys condition (Figure 1D) is a reflection of the decreased TCA cycle activity.
Fatty acid β-oxidation (FAO) can be an important part of energy metabolism in the cell, particularly when the TCA is limited; indeed, FAO can yield even larger amounts of ATP than the catabolism of glucose alone.[50] To generate energy, fatty acids need to undergo β-oxidation inside the mitochondria to produce NADH, FADH2 and acetyl-CoA. However, the lipids need to be transported into the mitochondria.[51] From the decreased abundance of acetylcarnitine as well as the lower expression of LACS and CPT1 (Figure 6J,I and Figure S6A, Supporting Information, respectively), we found that the fatty acids were neither activated by the addition of the CoA nor transported through the mitochondrial inner membrane in the low Cys condition. Other studies have shown that oxidative stress can inhibit PPARγ, which controls the activity of LACS; therefore, the redox imbalance present when the Cys level is limited is a potential cause for the decreased fatty acid activation.[52] Furthermore, although FAO can contribute a substantial amount of energy to the cell, if the cell does not have a functional TCA cycle and ETC, fatty acid β- oxidation cannot occur.[50] Additionally, an increased abundance of acetyl-coA and malonyl CoA is likely to occur due to the dysfunction of the TCA cycle. Malonyl CoA is known to inhibit CPT1, but the mechanism is not completely understood.[53] As a result of the oxidative stress due to the low Cys feed, the fatty acids were likely unable to provide the necessary cellular energy to maintain the high production of the mAb, leading to the dramatic loss in specific productivity and titer. As a further consequence, cells fed insufficient levels of Cys exhibited autophagy in which large amounts of lipid degradation products were released in the cytosol, especially lysophospholipids and glycerophosphate head-groups (Figure 6C–F). The high ROS condition in the cells could also contribute to phospholipid breakdown. This degradation is evidenced by the very large increase (≈80-fold) in released GPC and GPE, making these molecules potentially sensitive and useful bioprocess markers sensing the overall metabolic health of the cells during bioprocessing.
From our in-depth study of the biology of the high producing CHO cells under redox imbalance and other stresses, novel opportunities for genetic engineering and potential media additives are suggested for improvement in CHO cell lines productivity. For example, enhancing the expression level of the enzyme PC by genetic means could help ensure that sufficient pyruvate is converted to oxaloacetate in order to maintain and even enhance TCA cycle functionality.[54] Additionally, feeding TCA cycle intermediates can potentially help maintain the functionality of the cycle and increase productivity, similar to what others have shown.[55] Others have also found that modifying genes controlling lipid metabolism such as sterol regulatory element binding factor 1 (SREBF1) and stearoyl CoA desaturase 1 (SCD1) can enhance secretion, highlighting the importance of lipidomics in bioprocess development.[56]
Our study also suggested potential markers to follow the bioprocess during production of the mAb. To provide examples, we had supernatant samples analyzed for extracellular metabolites using GC-MS and LC-MS from the same culture days as the samples for ‘omics studies, as shown in Figure 7 and detailed in Supporting Information. GC-MS results revealed higher levels of extracellular Ala from the transamination reaction as early as day 6 for the low Cys fed level (Figure 7A). Additionally, using the GC-MS method, many extracellular sugars such as fructose were found to be enhanced (Figure 7B), mirroring the time course measurements of intracellular sugars. Further examples are provided in Figure S7, Supporting Information. In addition, LC-MS analysis showed quite high levels of GPC and GPE in the supernatant (Figure 7C,D) similar to the intracellular trends. As we have noted, these latter molecules have the potential to be quite sensitive markers.
Figure 7.
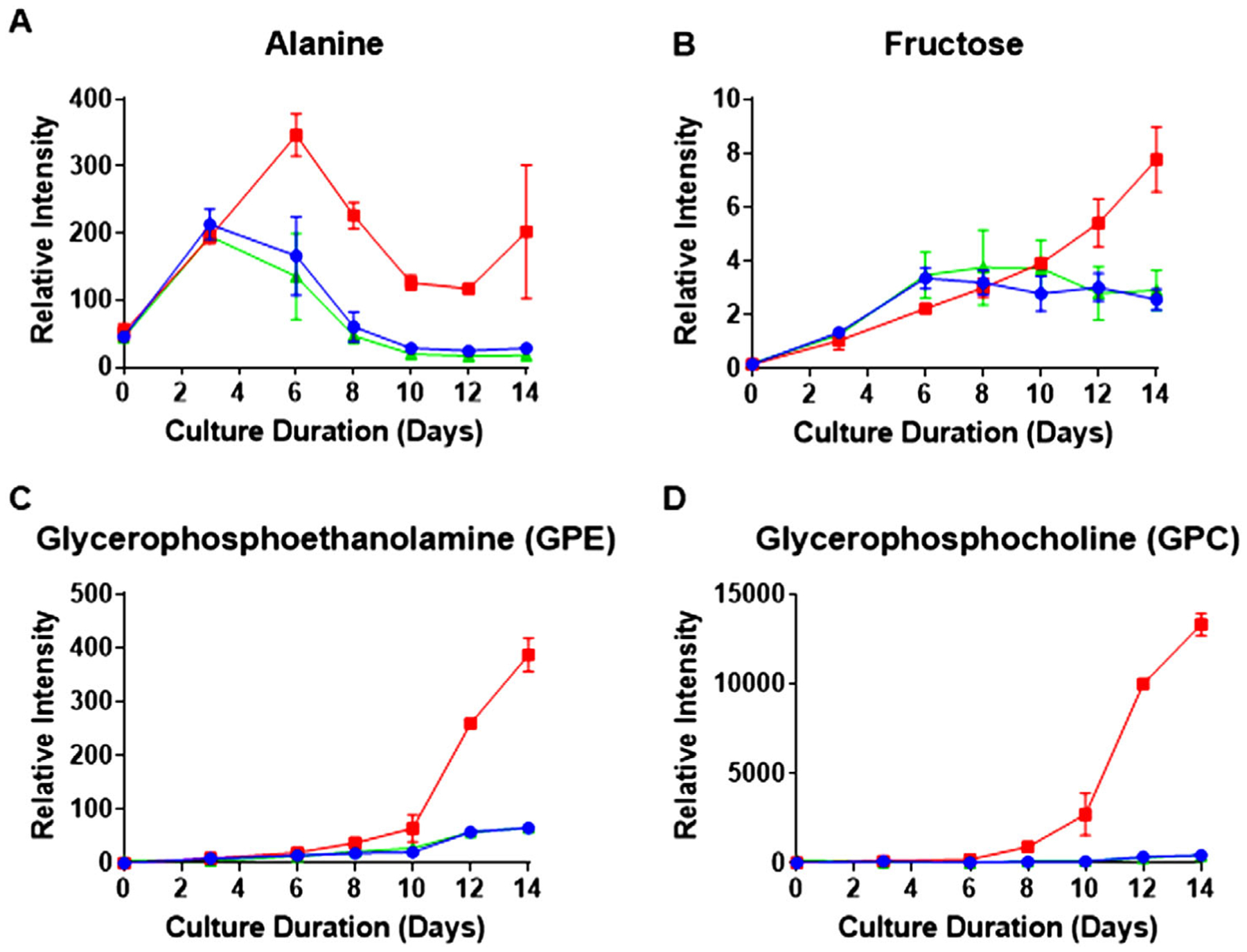
Extracellular metabolite time course profiles as a function of culture duration. Red, blue, and green represent the three Cys feed concentrations low, control, and high Cys, respectively. A,B) show the average time course of extracellular metabolites measured by GC-MS with error bars indicating standard deviation of relative intensity (n = 2). Both alanine and fructose are among the metabolites observed to increase in the supernatant. C,D) show the increase in lipid degradation products measured in the supernatant by LC-MS with error bars also indicating standard deviation of relative intensity (n = 2).
In conclusion, our analysis revealed the far-reaching impact of redox imbalance, ER stress, mitochondrial dysfunction, and amino acid limitation on various CHO metabolic pathways, ranging from lower energy generation to protein misfolding during mAb bioprocessing. By leveraging multi-omics analysis and using Cys feed level as a sensitive probe into metabolism and redox balance, we were able to achieve a deeper understanding of the CHO biotherapeutic production platform. Multi-omics analysis can thus help pave the way for improved efficiency of bioprocessing and aid in the manufacture of new effective therapies to alleviate patient disease.
4. Experimental Section
Cell Culture Conditions and Samples:
The experimental procedure for the 5 L fed-batch bioprocess was described previously.[10] Briefly, CHO DG44 cells expressing an IgG1 recombinant monoclonal antibody (mAb) were cultured for 14 days in nine bioreactors, consisting of three biological replicates of three different Cys feed concentrations, with daily feeding.[57] Bolus addition of chemically defined, proprietary media was fed once a day to the bioreactors after sampling while a separate Cys feed stock solution was used to deliberately reduce the Cys feed concertation from the control condition by 15% (referred to as low Cys) and increased by 20% (referred to as high Cys), as previously explained. Daily off-line measurements of pH, pCO2, and O2 were performed, prior to feeding, using a BGA500 (Siemens Healthcare, Tarrytown, NY) while a Cedex BioHT (Roche, Indianapolis, IN) was used to measure ammonia and lactate concentrations in the supernatant. Cell pellet samples were saved from all the Cys feed conditions for multi-omics analysis (transcriptomics, proteomics, metabolomics, and lipidomics) on days 0, 3, 6, 8, 10, 12, and 14 from the three biological replicates prior to bolus feeding. Similarly, supernatant samples were saved from the same days and filtered using a 0.2 μm PTFE filter (Millipore Inc., Billerica, MA) for extracellular analysis. Clarified cell culture supernatant samples from days 7, 10, 12, 13, and 14 from all replicates were saved and stored at −70 °C until mAb purification followed by N-glycosylation analysis. Details of the protein purification and glycosylation analysis are described in the Supporting Information.
Multi-Omics Analysis:
A cell culture sample containing 30 × 106 cells was saved from each 5 L bioreactor as described earlier for intracellular multi-omics analysis (transcriptomics, proteomics, metabolomics, and lipidomics). Here, a brief summary is provided of each of the ‘omics acquisition methodologies while the detailed information is presented in the Supporting Information. For transcriptomics analysis, similar to what has been presented previously,[10] RNA was extracted and purified using an RNA purification kit following the manufacturer’s protocol (Norgen Biotek Corp.). 500 ng of RNA were used for poly A library construction and sequenced on a HiSeq 2500 (Illumina Inc., San Diego CA).[10]
For proteomics analysis, 2D liquid chromatography-mass spectrometry (2D-LC-MS) was performed using tandem mass tag (TMT) methodology (Thermo Fisher Scientific) as previously.[10] Briefly, peptides were first separated on a high pH reversed-phase column (Zorbax 300 extend C18, 300 Å, 3.5 μm beads, 2.1 mm ID × 150 mm, Agilent Technologies, Santa Clara, CA) with collection of fractions at 2-min intervals using an automated fraction collector (Gilson Inc., Middleton, WI). In the second dimension, a low pH nanoflow reversed-phase separation was run on an Easy-Spray PepMap C18 column (75 μm × 500 mm, 100 Å, 2 μm) (Thermo Fisher Scientific) coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific). Resulting LC-MS raw files were subsequently processed using Proteome Discoverer 2.1 (Thermo Fisher Scientific) and searched against the NCBI CHO database using Sequest HT.[3]
Intracellular metabolomic samples were collected and washed with cold phosphate-buffered saline (PBS) using protocols developed by Metabolon Inc. (Durham, NC).[58] Metabolomics analysis was performed by Metabolon, following their standard protocols,[59] and results are reported as relative abundances, as described previously.[10] Extracellular metabolites were analyzed by both GC-MS and HILIC LC-MS, as detailed in the Supporting Information, to complement the intracellular results.
For intracellular lipidomics analysis, lipid sample preparation and extraction were performed similarly to what has been previously described.[60] Briefly, a double extraction procedure was performed using methyl tert-butyl ether (MTBE) and optima grade H2O to produce phase separation. Chromatographic separation of lipids was conducted on a charged surface hybrid (CSH) C18 column (2.1 mm I.D. × 150 mm, 130 Å,1.7 μm) (Waters Corp., Milford, MA)[61] and coupled to an LTQ-Orbitrap mass analyzer (Thermo Fisher Scientific) operating in the data-dependent acquisition mode. Positive and negative mode ESI spectra were acquired in separate injections of the same sample. Data processing was conducted with the software program Compound Discoverer 2.0 (Thermo Fisher Scientific), and identification was performed using a combined MS/MS lipid database from MS-Dial and GREAZY.[62,63] Details of the analysis approach and database generation have been described previously.[8]
Lipid nomenclature used throughout this paper follows the guidance of the LipidMAPS Consortium Standardized Notation.[64] Briefly, the first two letters indicate the lipid class while the number in the parenthesis indicates the number of carbons in the fatty acid chain followed by the number of double bonds present.
Multi-Omics Data Integration and Analysis:
For each of the ‘omics experiments, fold change ratios between the low Cys condition and control time course data were calculated for each culture day and analyzed. p- values were calculated using the two-tailed t-test with an unequal variance to determine the statistical significance of the comparison.[65] Numerical ratios greater than 1.2 and less than 0.8 with a p-value less than 0.05 were selected as minimum thresholds for difference between the two feed levels and were used for the color coding in the pathway figures. By comparing time course changes and trends, we were able to use multiple data points to strengthen our hypotheses. Additional proteomics results for proteins described in the pathway figures are provided in the Supporting Information.
Supplementary Material
Acknowledgements
The authors thank Normand Allaire, Ravi Challa, Joost Groot, and Chris Roberts, all of Biogen, for their assistance with the RNA sequencing. The authors also thank the West Coast Metabolomics Core for the extracellular GC-MS analysis. A.R.I. acknowledges support by the NIH awards 1R01GM120272 and R01CA218500.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/biot.201900565
Contributor Information
Rachel Chen, Analytical Development, Biogen Inc., Cambridge, MA 02142, USA.
Li Zang, Analytical Development, Biogen Inc., Cambridge, MA 02142, USA.
References
- [1].Dalakas MC, Nat. Clin. Pract. Neurol 2008, 4, 557. [DOI] [PubMed] [Google Scholar]
- [2].Baycin Hizal D, Tabb DL, Chaerkady R, Chen L, Lewis NE, Nagarajan H, Sarkaria V, Kumar A, Wolozny D, Colao J, Jacobson E,Tian Y, O’Meally RN, Krag SS, Cole RN, Palsson BØO, Zhang H, Betenbaugh M, Baycin-Hizal D, J. Proteome Res 2012, 11, 5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu Z, Dai S, Bones J, Ray S, Cha S, Karger BL, Li JJ, Wilson L,Hinckle G, Rossomando A, Biotechnol. Prog 2015, 31, 1026. [DOI] [PubMed] [Google Scholar]
- [4].Sellick CA, Croxford AS, Maqsood AR, Stephens GM, Westerhoff HV, Goodacre R, Dickson AJ, Biotechnol. J 2015, 10, 1434. [DOI] [PubMed] [Google Scholar]
- [5].Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ, Curr. Opin. Biotechnol 2013, 24, 1102. [DOI] [PubMed] [Google Scholar]
- [6].Gao Y, Ray S, Dai S, Ivanov AR, Abu-Absi NR, Lewis AM, Huang Z, Xing Z, Borys MC, Li ZJ, Karger BL, Biotechnol. J 2016, 11, 1190. [DOI] [PubMed] [Google Scholar]
- [7].Lewis AM, Abu-Absi NR, Borys MC, Li ZJ, Biotechnol. Bioeng 2016, 113, 26. [DOI] [PubMed] [Google Scholar]
- [8].Ali AS, Raju R, Ray S, Kshirsagar R, Gilbert A, Zang L, Karger BL, Biotechnol. J 2018, 13, 1700745. [DOI] [PubMed] [Google Scholar]
- [9].Cui X, Navneet S, Wang J, Roon P, Chen W, Xian M, Smith SB, Investig. Opthalmology Vis. Sci 2017, 58, 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ali AS, Raju R, Kshirsagar R, Ivanov AR, Gilbert A, Zang L, Karger BL, Biotechnol. J 2019, 14, 1800352. [DOI] [PubMed] [Google Scholar]
- [11].Stipanuk MH, Annu. Rev. Nutr 2004, 24, 539. [DOI] [PubMed] [Google Scholar]
- [12].Fan Y, Jimenez Del Val I, Müller C, Wagtberg Sen J, Rasmussen SK,Kontoravdi C, Weilguny D, Andersen MR, Biotechnol. Bioeng 2015, 112, 521. [DOI] [PubMed] [Google Scholar]
- [13].Templeton N, Dean J, Reddy P, Young JD, Biotechnol. Bioeng 2013, 110, 2013. [DOI] [PubMed] [Google Scholar]
- [14].Kilberg MS, Pan Y-X, Chen H, Leung-Pineda V, Annu. Rev. Nutr 2005, 25, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Méndez-Lucas A, Hyroššová P, Novellasdemunt L, Viñals F, Perales JC, J. Biol. Chem 2014, 289, 22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gawlitzek M, Ryll T, Lofgren J, Sliwkowski MB, Biotechnol. Bioeng 2000, 68, 637. [DOI] [PubMed] [Google Scholar]
- [17].Hong JK, Cho SM, Yoon SK, Appl. Microbiol. Biotechnol 2010, 88, 869. [DOI] [PubMed] [Google Scholar]
- [18].Cao SS, Kaufman RJ, Antioxid. Redox Signaling 2014, 21, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hetz C, Nat. Rev. Mol. Cell Biol 2012, 13, 89. [DOI] [PubMed] [Google Scholar]
- [20].Delic M, Rebnegger C, Wanka F, Puxbaum V, Haberhauer-Troyer C,Hann S, Köllensperger G, Mattanovich D, Gasser B, Free Radicals Biol. Med 2012, 52, 2000. [DOI] [PubMed] [Google Scholar]
- [21].Tu BP, Weissman JS, J. Cell Biol 2004, 164, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jadhav K, Zhang Y, Liver Res 2017, 1, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Senft D, Ronai ZA, Trends Biochem. Sci 2015, 40, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Capitanio D, Fania C, Torretta E, Vigano A, Moriggi M, Bravatà V,Caretti A, Levett DZH, Grocott MPW, Samaja M, Cerretelli P,Gelfi C, Sci. Rep 2017, 7, 9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hartley F, Walker T, Chung V, Morten K, Biotechnol. Bioeng 2018, 115, 1890. [DOI] [PubMed] [Google Scholar]
- [26].Deberardinis RJ, Cheng T, Oncogene 2010, 29, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang F, World J Biol. Chem 2012, 3, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guo Y, Cordes KR, V Farese R, Walther TC, J. Cell Sci 2009, 122, 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kohlwein SD, Petschnigg J, Curr. Hypertens. Rep 2007, 9, 455. [DOI] [PubMed] [Google Scholar]
- [30].Listenberger LL, Han X, Lewis SE, Cases S, V Farese R, Ory DS, Schaffer JE, Proc. Natl. Acad. Sci. U. S. A 2003, 100, 3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Walther TC, V Farese R, Annu. Rev. Biochem 2012, 81, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RGW, J. Biol. Chem 2004, 279, 3787. [DOI] [PubMed] [Google Scholar]
- [33].Evans AM, Fornasini G, Clin. Pharmacokinet 2003, 42, 941. [DOI] [PubMed] [Google Scholar]
- [34].Kerner J, Hoppel C, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2000, 1486, 1. [DOI] [PubMed] [Google Scholar]
- [35].Nyman LR, Cox KB, Hoppel CL, Kerner J, Barnoski BL, Hamm DA, Tian L, Schoeb TR, Wood PA, Mol. Genet. Metab 2005, 86, 179. [DOI] [PubMed] [Google Scholar]
- [36].Bhagavan NV, Ha C-E, Essentials of Medical Biochemistry, Elsevier, New York: 2011. [Google Scholar]
- [37].Tang Y, Zhou J, Hooi SC, Jiang YM, Lu GD, Oncol. Lett 2018, 16, 1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Orellana CA, Marcellin E, Schulz BL, Nouwens AS, Gray PP,Nielsen LK, J. Proteome Res 2015, 14, 609. [DOI] [PubMed] [Google Scholar]
- [39].Chakravarthi S, Bulleid NJ, J. Biol. Chem 2004, 279, 39872. [DOI] [PubMed] [Google Scholar]
- [40].Chong WPK, Thng SH, Hiu AP, Lee D-Y, Chan ECY, Ho YS, Biotechnol. Bioeng 2012, 109, 3103. [DOI] [PubMed] [Google Scholar]
- [41].He L, He T, Farrar S, Ji L, Liu T, Ma X, Cell. Physiol. Biochem 2017, 44, 532. [DOI] [PubMed] [Google Scholar]
- [42].Almanza A, Carlesso A, Chintha C, Creedican S, Doultsinos D, Leuzzi B, Luís A, McCarthy N, Montibeller L, More S, Papaioannou A,Püschel F, Sassano ML, Skoko J, Agostinis P, de Belleroche J, Eriksson LA, Fulda S, Gorman AM, Healy S, Kozlov A, Muñoz-Pinedo C, Rehm M, Chevet E, Samali A, J. FEBS 2019, 286, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Valera-Alberni M, Canto C, Cell Stress 2018, 2, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Min Lee G, Koo J, Encyclopedia of Industrial Biotechnology, John Wiley & Sons, Hoboken, NJ: 2010. [Google Scholar]
- [45].Pan Y-X, Chen H, Thiaville MM, Kilberg MS, Biochem. J 2007, 401, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Duarte TM, Carinhas N, Barreiro LC, Carrondo MJT, Alves PM, Teixeira AP, Biotechnol. Bioeng 2014, 111, 2095. [DOI] [PubMed] [Google Scholar]
- [47].Ghaffari N, Jardon MA, Krahn N, Butler M, Kennard M, Turner RFB, Gopaluni B, Piret JM, Biotechnol. Prog 2019, 10, e2946. [DOI] [PubMed] [Google Scholar]
- [48].Montal ED, Dewi R, Bhalla K, Ou L, Hwang BJ, Ropell AE, Gordon C, Liu WJ, DeBerardinis RJ, Sudderth J, Twaddel W, Boros LG, Shroyer KR, Duraisamy S, Drapkin R, Powers RS, Rohde JM, Boxer MB, Wong KK, Girnun GD, Mol. Cell 2015, 60, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Vincent EE, Sergushichev A, Griss T, Gingras MC, Samborska B,Ntimbane T, Coelho PP, Blagih J, Raissi TC, Choinière L, Bridon G, Loginicheva E, Flynn BR, Thomas EC, Tavaré JM, Avizonis D, Pause A, Elder DJE, Artyomov MN, Jones RG, Mol. Cell 2015, 60, 195. [DOI] [PubMed] [Google Scholar]
- [50].Houten SM, Wanders RJA, Inherited Metab J. Dis 2010, 33, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Giudetti AM, Stanca E, Siculella L, Gnoni GV, Damiano F, Int. J. Mol. Sci 2016, 17, 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Blanquicett C, Kang B-Y, Ritzenthaler JD, Jones DP, Hart CM, Free Radicals Biol. Med 2010, 48, 1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Qu Q, Zeng F, Liu X, Wang QJ, Deng F, Cell Death Dis 2016, 7, e2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim SH, Lee GM, Appl. Microbiol. Biotechnol 2007, 76, 659. [DOI] [PubMed] [Google Scholar]
- [55].Zhang X, Jiang R, Lin H, Xu S, Biotechnol. Prog 2020, 3, e2975. [DOI] [PubMed] [Google Scholar]
- [56].Budge JD, Knight TJ, Povey J, Roobol J, Brown IR, Singh G, Dean A, Turner S, Jaques CM, Young RJ, Racher AJ, Smales CM, Metab. Eng 2020, 57, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang WC, Lu J, Kwiatkowski C, Yuan H, Kshirsagar R, Ryll T, Huang Y-M, Biotechnol. Prog 2014, 30, 616. [DOI] [PubMed] [Google Scholar]
- [58].Luo J, Vijayasankaran N, Autsen J, Santuray R, Hudson T, Amanullah A, Li F, Biotechnol. Bioeng 2012, 109, 146. [DOI] [PubMed] [Google Scholar]
- [59].Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA, V Milburn M, Pharma-cogenomics 2008, 9, 383. [DOI] [PubMed] [Google Scholar]
- [60].Matyash V, Liebisch G, V Kurzchalia T, Shevchenko A, Schwudke D, J. Lipid Res 2008, 49, 1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bird SS, Marur VR, Stavrovskaya IG, Kristal BS, Anal. Chem 2012, 84, 5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M, Nat. Methods 2015, 12, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kochen MA, Chambers MC, Holman JD, Nesvizhskii AI, Weintraub ST, Belisle JT, Islam MN, Griss J, Tabb DL, Anal. Chem 2016, 88, 5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ,Schmitz G, Spener F, Wakelam MJO, J. Lipid Res 2013, 54, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, Mclean JA,J. Am. Soc. Mass Spectrom 2016, 27, 1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


