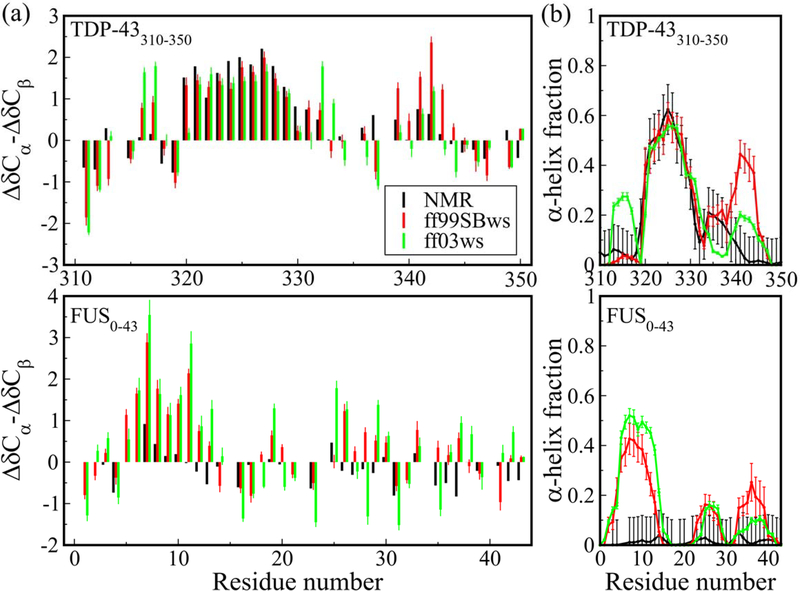
Figure 1.
(a) Chemical shifts ΔδCα − ΔδCβ and (b) α-helix propensities of TDP-43310−350 and FUS0−43 using all-atom force fields ff99SBws and ff03ws, compared to experimental results. Chemical shifts are calculated from protein structure using SPARTA+ algorithm. Secondary structures are assigned by DSSP. Experimental values of secondary structure propensities are calculated from NMR chemical shifts using the δ2D program.