Fig. 5. Mitovesicles are metabolically active EVs with a specific subset of mitochondrial proteins.
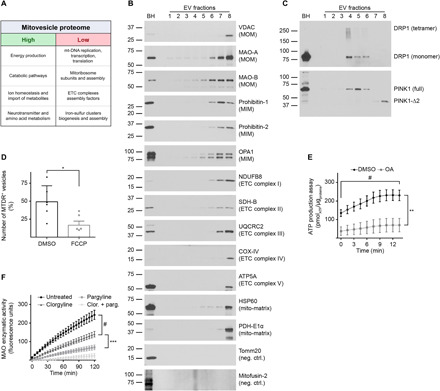
(A) EVs were isolated from brains of 12-month-old wild-type mice and fractionated through an OptiPrep step gradient. Fr8 EV lysates were analyzed by LC-MS, and the mitochondrial proteins found were clustered according to their known functions. Mitochondrial pathways that were highly underrepresented are indicated as “high” and “low,” respectively. (B) Representative Western blot analysis of brain EVs with antibodies to mitochondrial proteins. (C) Representative Western blot analyses of brain EVs using antibodies to DRP1 and PINK1. (D) Relative number of MTDR-positive (MTDR+) vesicles found in Fr8 EVs as quantified by NTA and expressed as percentage over the total number of EVs. As a control, samples were treated with the uncoupler FCCP that dissipates mitochondrial potential. (E) Analysis of ATP levels in brain Fr8 using a luciferase-based assay. As a control, samples were treated with oligomycin + antimycin-A (OA). Each experiment was run in triplicate. Chemiluminescence was quantified every 1.5 min. (F) MAO activity was measured in Fr8 brain EVs using a commercial MAO enzymatic activity assay. As controls, samples were treated with clorgyline (MAO-A inhibitor), pargyline (MAO-B inhibitor), or both. Fluorescence was read every 5 min. For additional data see table S2. Brain homogenate (BH) was loaded as a reference. Statistical test in (D): Student’s t test; (E) and (F): two-way ANOVA with Tukey’s multiple comparisons test. Number of independent isolations: (A), 3; (B) and (C), 10; (D), 6; (E) and (F), 4. Data shown in (D) to (F): Means ± SD. *P < 0.05, **P < 0.01, ***P < 10−3, and #P < 10−4.
