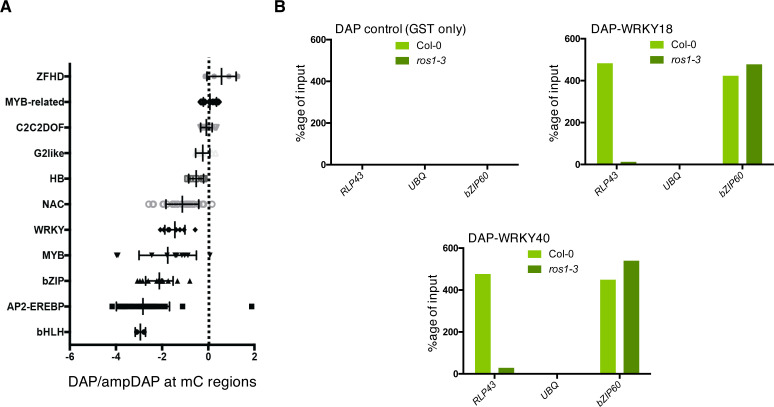
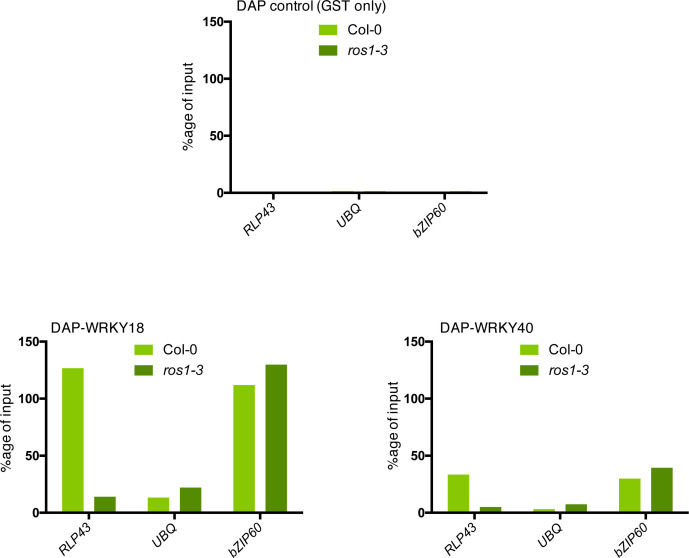
Figure 5. ROS1-directed demethylation is crucial for the DNA binding of WRKY18 and WRKY40 at the RLP43 promoter.
(A) DAP-seq data from O'Malley et al., 2016 showing that WRKY family members are generally sensitive to DNA methylation. Graph representing ratio of binding capacity in DAP versus ampDAP data at regions methylated in Col-0 and for the different transcription factor family members used in this study. (B) The ability of WRKY18 and WRKY40 to bind DNA corresponding to the demethylated region of the RLP43 promoter is abolished in the ros1-3 mutant background. DAP-qPCR analysis at the RLP43 promoter upon pull-down of Col-0 or ros1-3 genomic DNA by GST (negative control), WRKY18-GST or WRKY40-GST. UBQ and bZIP60 served as negative and positive controls, respectively.