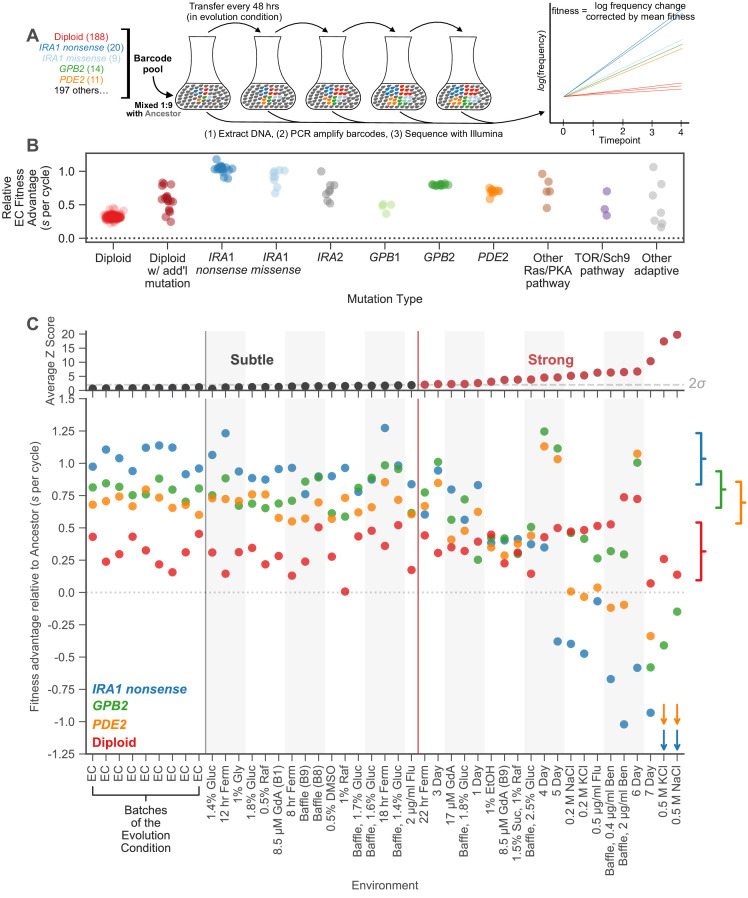
Figure 2. Measuring fitness for a collection of adaptive mutants across many environments reveals gene-by-environment interactions.
(A) Schematic of fitness measurement procedure. Adaptive mutants tagged with DNA barcodes are pooled at a 1:9 ratio with an ancestral reference strain. The pool is then propagated for several growth cycles, where the population is diluted into fresh media at fixed time intervals. DNA is extracted from each time-point, and the barcode region is PCR amplified and then sequenced. A mutant’s relative fitness is calculated based on the rate of change of its barcode’s frequency, corrected for the mean fitness of the population (see Materials and methods). Relative fitness is calculated in units of ‘per cycle’, representing the improvement of each barcode relative to the reference over the course of the time between transfers. (B) Fitness advantage of each mutant in the evolution condition relative to the ancestor. This fitness advantage is measured per transfer cycle and calculated as the average across all nine Evolution Condition (EC) batches. (C) (top) Environments are ordered from left to right depending on the degree to which they perturb mutant fitness from the average fitness observed across all EC batches. Environments in which average mutant fitness is within two standard deviations of average mutant fitness across EC batches are denoted in black and make up the subtle perturbation set. Environments in which aggregate mutant behavior exceeds two standard deviations are shown in red and make up the strong perturbations set. (bottom) This plot displays, for the four most common types of adaptive mutation observed in response to glucose limitation (Venkataram et al., 2016a), the average fitness in each of the 45 environments we study. Brackets on the right represent the amount of variation in fitness observed for each type of mutation across the EC batches, with the notch representing the mean and the arms representing two standard deviations on either side of the mean. For visualization purposes, we represent relative fitness values below −1.25 as arrows. Specifically, PDE2 mutants (orange arrows) have on average fitness −3.3 and −3.4 in 0.5 M KCl and 0.5 M NaCl, respectively. IRA1 nonsense mutants (blue arrows) have an average fitness −3.0 and −4.2 in 0.5 M KCl and 0.5 M NaCl, respectively.
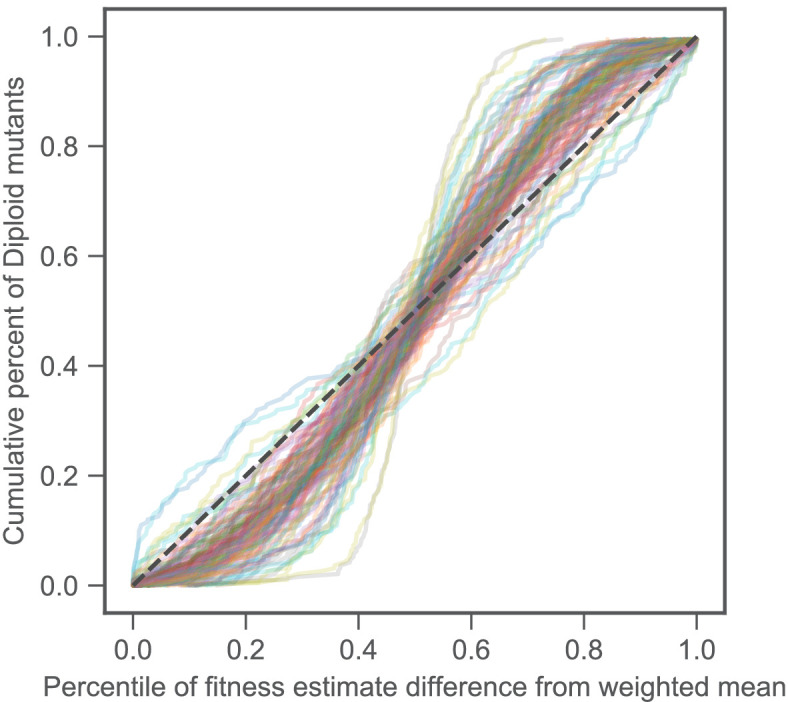
Figure 2—figure supplement 1. Noise model is a conservative measure of uncertainty.