Figure 2. A biochemically-derived model for a DNA N6mA role in nucleotide excision repair.
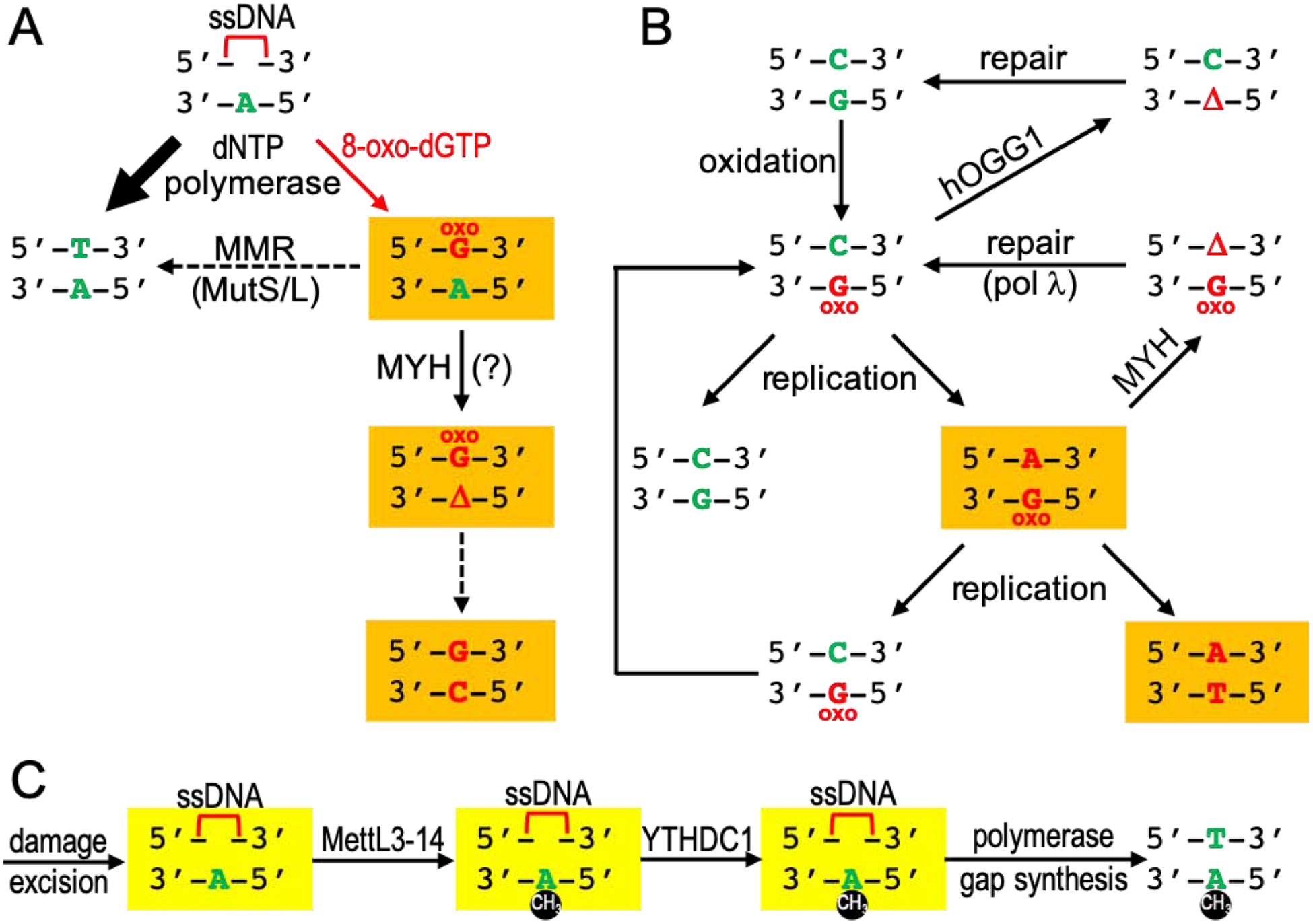
(A) The red bracket represents an ssDNA gap (~25–30 nt) after nucleotide excision (top). DNA synthesis would be largely accurate because T is preferentially inserted opposite A (left bold arrow). However, reactive oxygen species can lead to the formation of 8-oxo-dGTP in the dNTP pool, and in the event that 8-oxo-dGTP is incorporated into DNA during synthesis, it may be either correctly base-paired opposite C (see Figure 1C) or mispaired opposite Ade (right red arrow; see Figure 1A). In the latter event, MMR most likely excises incorporated 8-oxoG from newly synthesized daughter DNA (dashed line) or base excision of the (correct) Ade by the MYH DNA glycosylase can result in a T→G transversion mutation (orange boxes). (B) Oxidation can occur directly to a G:C base pair already in the DNA, resulting in an 8-oxoG:C pair. The adduct can be excised from DNA by hOGG1 glycosylase, and subsequently repaired via base excision repair (BER) to restore the original G:C base pair (top right). If the 8-oxoG adduct is not removed prior to DNA replication, DNA synthesis may retain the 8-oxoG and form an 8-oxoG:A mispair (middle right, orange box; see also Figure 1D–F). The misincorporated Ade can be removed by the MYH DNA glycosylase and replaced by C in the single-nucleotide gap via polymerase λ, yielding a further chance for removal of the 8-oxoG lesion by hOGG1. If not repaired in time, a second round of replication can yield a C→A transversion mutation (orange box, bottom right). (C) A model of N6mA reducing misincorporation of 8-oxoG opposite to Ade. Between the nucleotide excision and DNA synthesis, a transient ssDNA gap (~25–30 nt; the red bracket) can become methylated by MettL3–14 and protected by YTHDC1.
