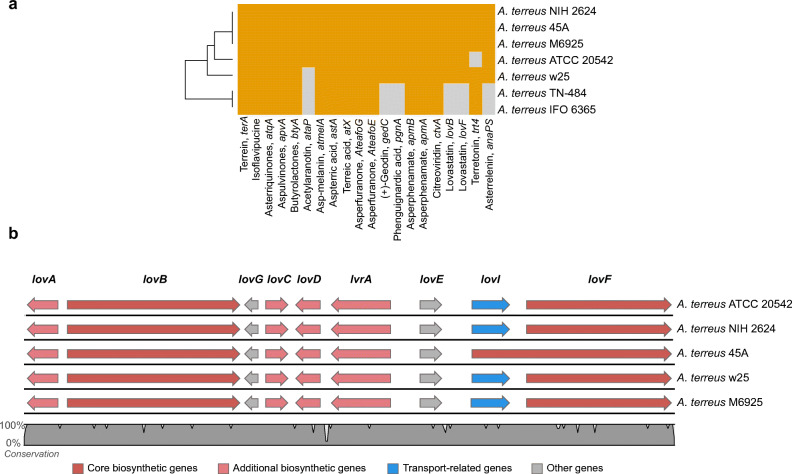
Fig. 4.
Comparison of the characterized secondary metabolism gene clusters in A. terreus strains. a Clustering of the A. terreus strains based on presence (blue tiles)/absence (gray tiles) of orthologs of A. terreus NIH 2624 core biosynthetic proteins involved in secondary metabolism, which were experimentally linked to their metabolic products in previous studies (as reviewed by Romsdahl and Wang 2019). Dendrogram was generated based on hierarchical clustering analysis. Y-axis: strain clustering; x-axis: protein clustering (dendrogram not shown). A. terreus T3 Kankrej was not included in the analysis due to unsatisfactory quality of its genome (high sequence fragmentation). b Comparison of lovastatin biosynthesis gene cluster conserved in five A. terreus genomes (gene names according to Dietrich and Vederas 2014)