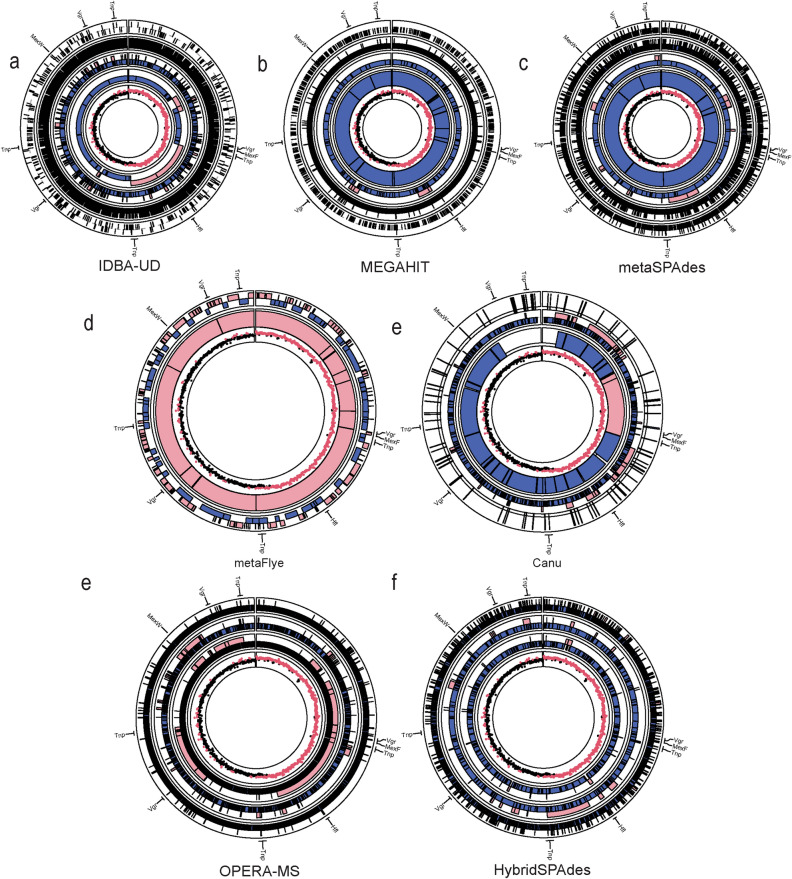
Figure 5.
Circular depictions of the Marinobacter hydrocarbonoclasticus ATCC 49,840 genome with assembler type from USA-AS samples. (a) MEGAHIT with 1 × , 5 × , 10 × , 50×, alternating between misassemblies (pink) and correct assemblies (blue). (b) metaSPAdes with 1 × , 5 × , 10 × , 50×. (c) IDBA-UD with 1 × , 5 × , 10 × , 50×. (d) metaFlye with 0.1 × , 1 × , and 5 × coverage. (d) Canu with 1 × and 5 × coverage (no Flye contigs mapped at 0.1 × coverage). (e) Canu with 0.1 × , 1 × , and 5 × coverage. (d) Canu with 1 × and 5 × coverage (no Flye contigs mapped at 0.1 × coverage). (f) OPERA-MS with 10 × short-read and 0.1 × , 1 × , and 5 × long-read coverage. (g) HybridSpades with 10 × short-read and 0.1 × , 1 × , and 5 × long-read coverage. Labels are regions of the genome annotated with ARGs or MGE genes. Internal red/black dots are GC skew of 1000 basepair sections of the genome (red is positive GC skew while red–black is negative GC skew). This figure was generated using the Circlize(v0.4.11) package in R(v3.5.0).