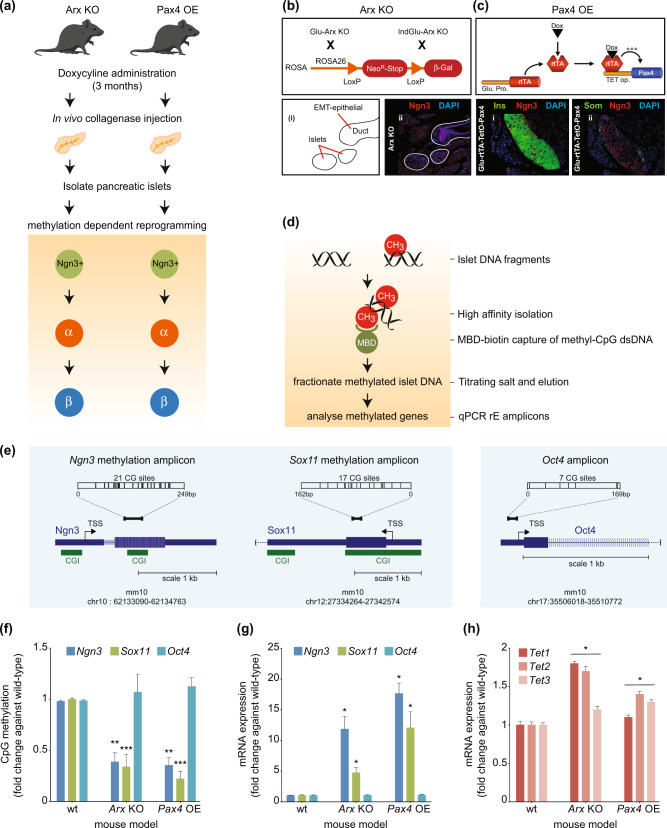
Fig. 1. DNA methylation-dependent reprogramming of islet cells derived from Arx knockout and overexpression of Pax4 animal models.
a Glu-rtTA::TetO-Pax4 animals were generated as previously described [7], treated with Dox at 4 weeks of age for 3 months. The ArxKO mouse line was crossed with Glu-rtTA transgenic line permitting the inducible deletion of Arx in adult α-cells [8]. Mice were then treated with Dox for 3 months. Islets from both transgenic lines were then purified to assess for methylation-dependent reprogramming. b, c Analysis of Ngn3 re-expression in the mouse pancreata. The expression of Ngn3 was analysed by immunohistochemistry in WT/Dox- controls and Dox-treated Pax4OE and ArxKO pancreata. Ngn3 labelling was absent in controls, while strongly re-expressed in induced animals (b, c). Ngn3 is re-expressed in the ductal lining and epithelium (b i and ii) as well as in the islets (b, c) of transgenic mice while being absent in controls. d Workflow of DNA methylation capture and analysis of transgenic mice islets using methyl-domain-binding (MBD) capture and downstream qPCR (MBD-qPCR) were used for the assessment of the reprogramming (rE) amplicons Ngn3 and Sox11. e The reprogramming amplicons Ngn3 and Sox11 were designed from the mouse genome assembly (mm10) using UCSC browser. CpG Islands (CGI) are shown in green for Ngn3 and Sox11. Oct4 does not have a CGI and served as a control for DNA methylation. The positions of the transcription start sites (TSS) are also shown relative to 1 kb scale. Chromosome positions are shown Ngn (chr10), Sox11 (chr12) and Oct4 (chr17). f DNA methylation analysis of islet development genes in transgenic mouse models, PaxOE and ArxKO. Data show DNA methylation (fold-change) for Ngn3 and Sox11. A member of the POU transcription factor family, Oct4, central to the machinery governing pluripotency served as an endogenous control and remains stable for DNA methylation. Error bars are defined as standard error of the mean (s.e.m) with significance calculated by comparing wild-type (wt) to transgenic mouse models PaxOE and ArxKO, using t-test (*P < 0.05, **P < 0.01, ***P < 0.001). g mRNA expression of genes associated with islet lineage reprogramming. Data shows gene expression (fold-change) normalised to housekeeping gene (H3F3A). SEM error bars with significance calculated by comparing wild-type (wt) to transgenic mouse models PaxOE and ArxKO, (*P < 0.05). h mRNA expression of ten eleven translocation (Tet) enzymes in transgenic mice islets. Error bars are defined as standard error of the mean (s.e.m) with significance calculated comparing wild-type (wt) to transgenic mouse models PaxOE and ArxKO, (*P < 0.05).