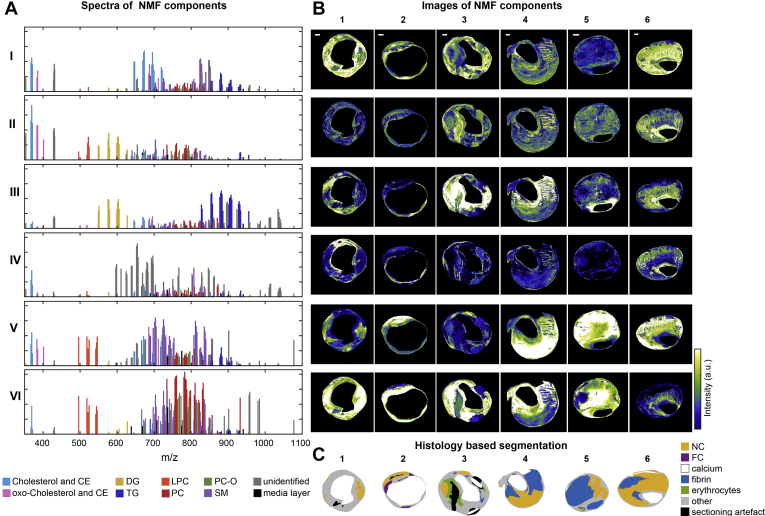
Fig. 4.
Unsupervised NMF of the 194 m/z-containing MALDI-MSI data. A: NMF spectra of the components showing the weight of each m/z value to that NMF component. M/z values in the NMF component spectra are labeled according to their assigned lipid class, showing a clear separation of lipid classes by means of NMF. B: Corresponding NMF-weighted images of the representative sections, showing the spatial distribution and relative intensities of the NMF components. C: Corresponding histology-based segmentation suggesting colocalization between certain NMF components and histologically relevant features. Scale bars are 1 mm. I–VI, NMF components; 1–6, representative tissue sections (same sections as depicted in Fig. 1); CE, cholesterol ester; DG, diacylglycerol; FC, foam cells; LPC, lysophosphatidylcholines; NC, necrotic core; NMF, nonnegative matrix factorization; PC, phosphatidylcholines; SM, sphingomyelin; TG, triacylglycerol.