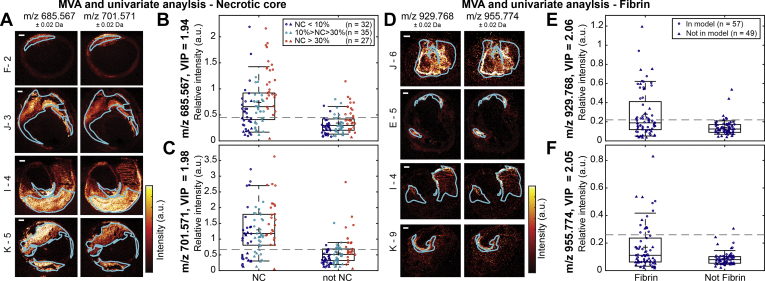
Fig. 5.
Univariate analysis comparing intensities in necrotic core (NC) versus not-NC areas and fibrin versus not-fibrin areas for two lipids. A: MALDI-MSI images of m/z 685.567 (oxoODE-CE [M+Na]+) and m/z 701.571 (SM(34:2) [M+H]+) of four different tissue sections with superimposed outlines of the NC segmentation. Boxplots of m/z 685.567 (B) and m/z 701.571 (C) showing the mean intensity of the pixels in the NC area and in the not-NC area. Datapoints are colored according to the size of NC present in the corresponding tissue section. D: MALDI-MSI images of m/z 929.768 (TG(58:9) [M+H]+ or TG(56:6) [M+Na]+) and m/z 955.774 (TG(60:10) [M+H]+ or TG(58:7) [M+Na]+) of four different tissue sections with the outlines of the fibrin segmentation. Boxplots of m/z 929.768 (E) and m/z 955.774 (F) showing the mean intensity of the pixels in the fibrin area and in the not-fibrin area. Only tissue sections containing NC or fibrin were included in these analyses. The dotted lines depict the maximum Youden's index, representing the optimal cutoff intensity value above which the m/z value is more likely to be associated with NC or fibrin. For m/z 685.567 the threshold is 0.45 and for m/z 701.571 it is 0.67, for m/z 929.768 it is 0.22 and for m/z 955.774 it is 0.26. All m/z values showed significantly different intensities between NC and not-NC or between fibrin and not-fibrin (P-value < 0.001). ○, data of the patients included in the multivariate model; Δ, data of patients that did not fit the model. Scale bars are 1 mm. Sections F-2 and I-4 are the same sections as 2 and 4 in Figs. 1 and 4, respectively. Sections J-6 and K-9 are also depicted in Fig. 3. MVA, multivariate analysis; NC, necrotic core; VIP, variable influence for projection.