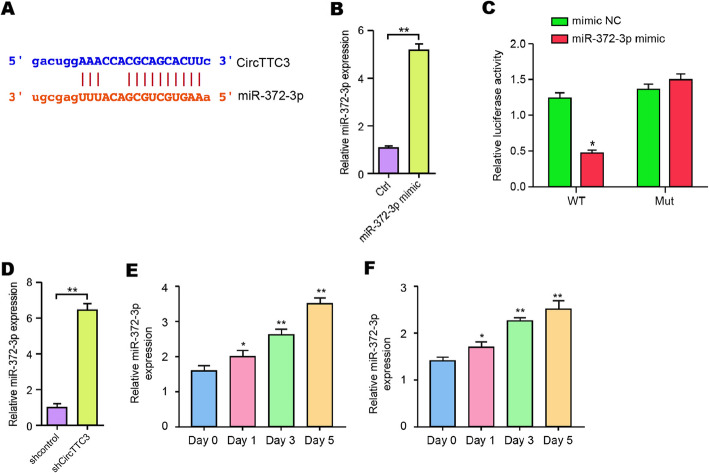
Fig. 4.
CircTTC3 is able to sponge miR-372-3p in NSCs. a The potential interaction between circTTC3 and miR-372-3p was identified by the bioinformatic analysis using ENCORI (http://starbase.sysu.edu.cn/index.php). b, c The NSCs were treated with the miR-372-3p mimic or control mimic. b The expression levels of miR-372-3p were measured by qPCR in the cells. c The luciferase activities of wild-type circTTC3 (WT) and circTTC3 with the miR-372-3p-binding site mutant (MUT) were determined by luciferase reporter gene assays in the cells. d The NSC cells were infected with lentiviral plasmids carrying circTTC3 shRNA or corresponding control shRNA. The expression of miR-372-3p was analyzed by qPCR in the cells. e, f The expression of miR-372-3p was measured by qPCR during NSC differentiation into astrocytes and neurons, respectively. Data are presented as mean ± SD. Statistic significant differences were indicated: *P < 0.05, **P < 0.01