Figure 1.
hPSC-CMs Express SARS-CoV-2 Receptors and Processing Factors
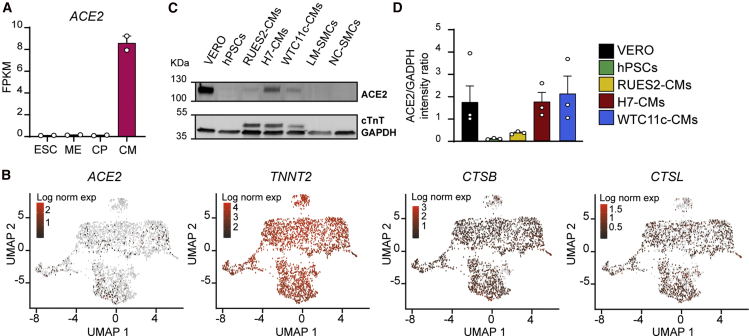
(A) RNA-seq during RUES2 hESC-CM differentiation. ACE2 is quantified as fragments per kilobase of transcript per million mapped reads (FPKM). Mean ± SEM of two independent experiments. ESC, embryonic stem cells (day 0); ME, mesoderm (day 2); CP, cardiac progenitors (day 5); CM, cardiomyocytes (day 14).
(B) Single-cell RNA-seq gene expression heatmaps from RUES2 hESC-CMs after dimensionality reduction through Uniform Manifold Approximation and Projection (UMAP). TNNT2 provides a pan-cardiomyocyte marker.
(C) Western blot for ACE2 in hPSC-CMs from multiple lines and different types of hPSC-SMCs. LM, lateral plate mesoderm-derived; NC, neural crest-derived; hPSCs, negative control; VERO cells, positive control.
(D) Quantification of ACE2 level normalized by GADPH. Mean ± SEM of three independent experiments.