Figure 1.
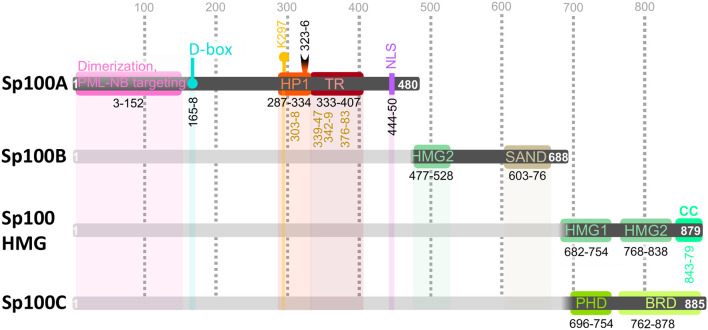
Sp100 isoform domain composition. Sp100A/B/C/high mobility group (HMG) share domain architecture within their first 477 amino acid (aa) residues: dimerization and promyelocytic leukemia-nuclear body (PML-NB) targeting (aa 3–152, pink), destruction-box (D-box, aa 165–168, teal); HP1 interacting region (aa 287–334, orange) encompassing a SUMO consensus motif (SCM) with Lys297 SUMO modification (K297, yellow pin) and SUMO-interacting motif (SIM, aa 323–326, black half-moon); trans-activating region (TR, aa 333–407, cherry); autoepitopes are indicated with vertical numbers in ochre below HP1 and TR segments (see EBV section); nuclear localization sequence (NLS, aa 440–450, purple). Sp100B/C/HMG are identical up to aa 685, which includes the high mobility group (HMG) 2 (aa 477–528, fern) and Sp100, AIRE-1, NucP41/75, DEAF-1 (SAND, aa 603-676, sand) DNA-binding domains. Sp100HMG contains two additional HMG (HMG1, aa 682–754; HMG2, aa 768–838) domains and a coiled coil (CC, aa 843–879, mint) domain. Sp100C contains a plant homeodomain (PHD, aa 696-754, green) and bromodomain (BRD, aa 762–878, light green) domain. C-terminal domain features are described in Table 1. Numbers indicate the positions of aa with each isoform. UniProt IDs: Sp100A, P23497-2; Sp100B, P23497-3; Sp100C, P23497-4; Sp100HMG, P23497-1. Further details on the Sp100 gene locus (ENSG00000067066) can be found at ENSEMBL1.