FIGURE 1.
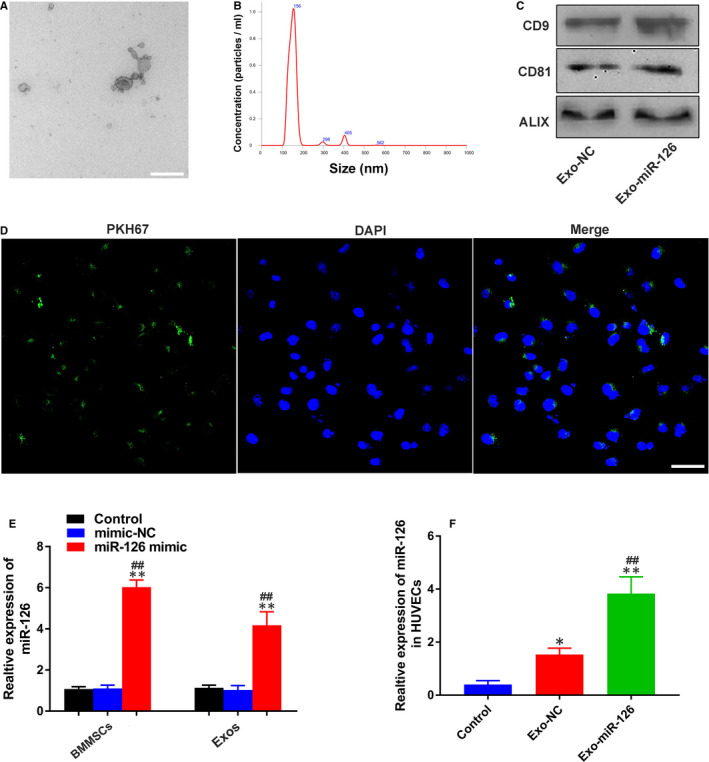
Identification and internalization of exosomes. (A) Morphology of exosomes under transmission electron microscopy (scale bar: 100 nm). (B) Size of exosomes identified by nanoparticle tracking analysis. (C) Exosomal surface markers CD9, CD81 and ALIX detected by Western blotting. (D) Fluorescence microscopy analysis of PKH67‐labelled exosomes (green) internalized by HUVECs. Nuclei were stained with DAPI (blue) for counterstaining (scale bar: 50 μm). (E) miR‐126 expression in BMMSCs and BMMSC‐derived exosomes detected by qRT‐PCR (n = 3), **P < 0.01 vs. Control group, ## P < 0.01 vs. mimic‐NC group (F) miR‐126 expression in HUVECs treated with PBS, Exo‐NC and Exo‐miR‐126 detected by qRT‐PCR (n = 3). *P < 0.05, **P < 0.01 vs. Control group, ## P < 0.01 vs. Exo‐NC group. HUVECs, human umbilical vein endothelial cells; Exo‐NC, exosomes derived from BMMSCs transfected with NC‐mimic; Exo‐miR‐126, exosomes derived from microRNA‐126 overexpressing bone marrow mesenchymal stem cells
