FIGURE 7.
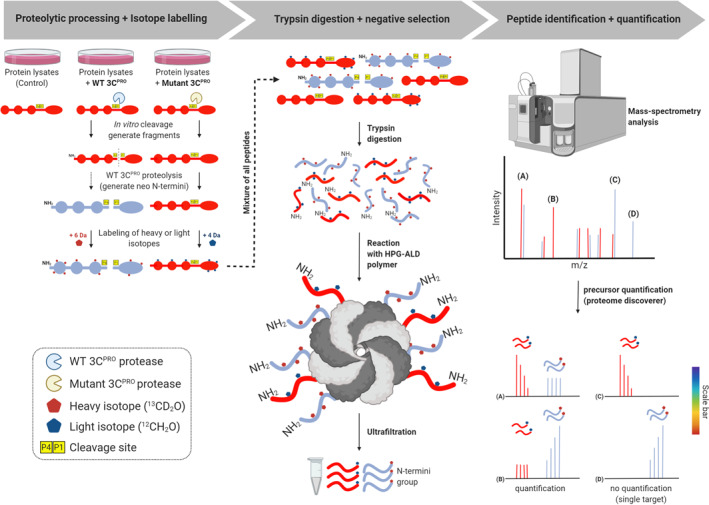
Schematic workflow of TAILS N‐terminomics screening of 3Cpro and 3CLprosubstrates. Schematic diagram depicts the TAILS workflow and scheme for identification of 3Cpro or 3CLpro substrates. In brief, protein samples from whole cell lysates were subjected to in vitro cleavage by either recombinant purified WT 3Cpro/3CLpro or mutant (C147A) 3Cpro/(C145A) 3CLpro, followed by N‐terminal enrichment using TAILS (left panel). Samples were then combined and subjected to pre‐TAILS shotgun‐like mass‐spectrometry analysis after complete digestion with trypsin. The exposed amine groups of N‐termini generated by the trypsin digestion were then removed by covalently coupling to a high‐molecular weight polyaldehyde polyglycerol polymer. This process allowed for selection via negative enrichment of blocked N termini (middle panel). Peptides were subsequently identified and quantified using high‐resolution mass spectrometry (indicated in the right panel). The resultant high‐confidence candidate substrates were determined through the analysis of the quantified heavy/light (H/L) ratio of dimethylation‐labeled semitryptic neo‐N terminus peptides. They will be subjected to further validation through similar in vitro cleavage assay by 3Cpro/3CLpro, followed by immunoblotting using specific antibodies; TAILS, terminal amine isotopic labeling of substrates; 3Cpro, 3C protease; 3CLpro, 3C‐like protease
