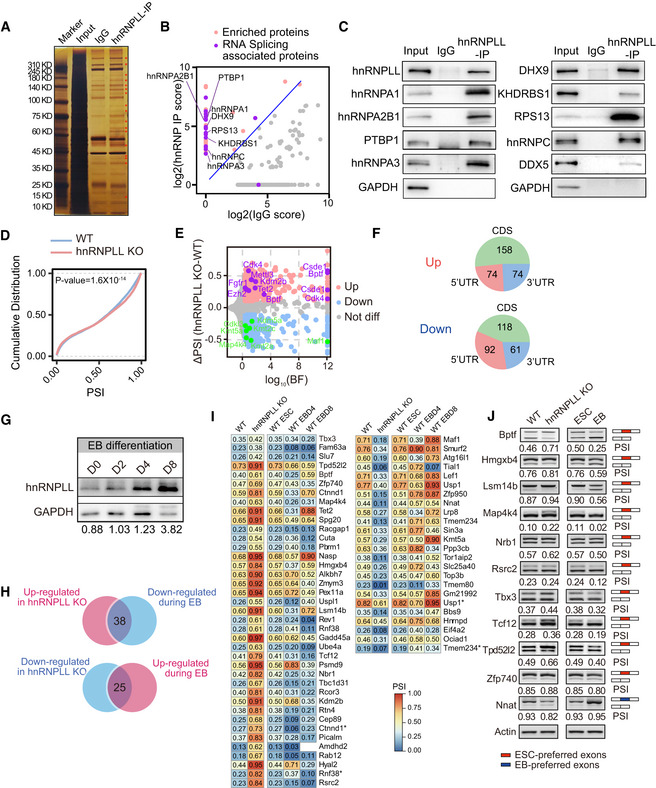
Silver nitrate‐stained NuPAGE gel of hnRNPLL‐interacting proteins. Red star indicate hnRNPLL‐specific interacting proteins compared with IgG.
Mass spectrum identification of the hnRNPLL interactome in ES cells. RNA splicing associated proteins are well enriched by hnRNPLL IP. The enrichment score was log2‐transformed before plotting.
Western blot validation the interactions between hnRNPLL and the indicated splicing‐associated proteins.
Cumulative distribution of PSI with exon skipping events between WT and hnRNPLL‐KO ES cells. Kolmogorov–Smirnov test was used to determine the significance.
Volcano plot of differential exon skipping events between WT and hnRNPLL‐KO ES cells. Indicated AS events were labeled in purple (up‐regulated) and green (down‐regulated) with gene names.
The regional distribution of differential exon skipping events between WT and hnRNPLL‐KO ES cells.
Western blot analysis of hnRNPLL protein during EB formation of WT ES cells at indicated days.
Venn plot showed the divergent exon skipping events between EB differentiation and hnRNPLL deletion. 38 exon skipping events that were inhibited by hnRNPLL but increased during EB differentiation, and 25 events that showed the opposite profile.
Heatmap of the 38 and 25 divergent exon skipping events in Fig
5I. * indicates the different exon skipping events of the same gene.
RT–PCR validation of partial divergent exon skipping events shown in Fig
5J. Red boxes represent ESC preferred exons, blue boxes indicate EB preferred exons.