Figure 8. PIDD1 localization to DAs is required for PIDDosome activation in response to DNA damage.
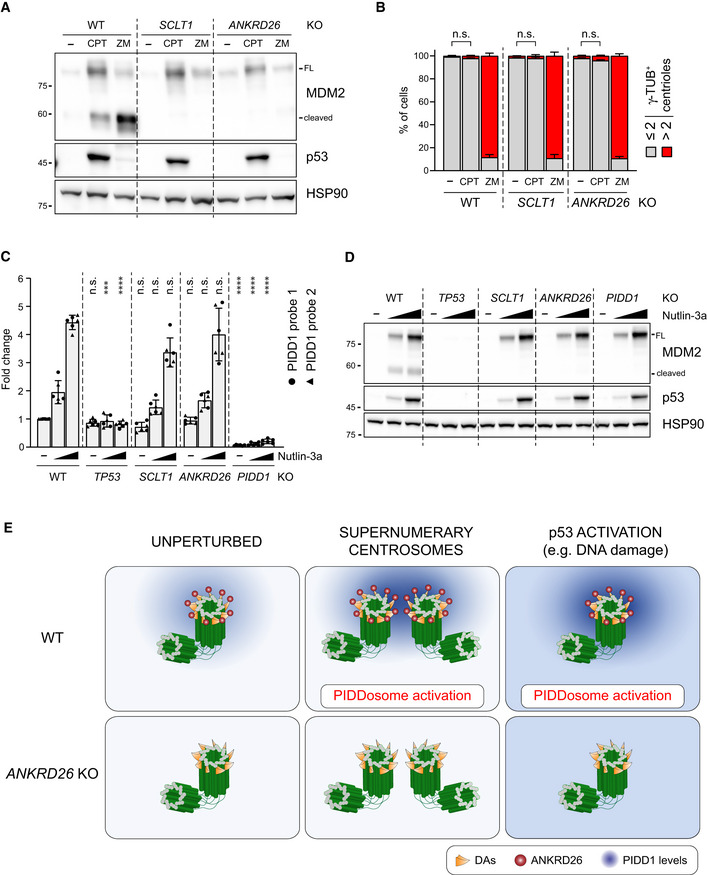
- A549 cells of the indicated genotypes were treated for 24 h as indicated (CPT = camptothecin; ZM = ZM447439). Samples were subjected to immunoblotting; n = 3 independent experiments.
- A549 cells treated as in (A) were subjected to fluorescence microscopy and centrosome abundance was assessed by visually scoring γ‐tubulin‐positive centrioles per cell. Mean values ± s.e.m. are reported. N = 3, ≥ 50 cells from each independent experiment. ANOVA test (n.s. = non‐significant).
- RT–qPCR analysis of PIDD1 mRNA expression in A549 cells upon treatment with increasing doses of Nutlin‐3a (i.e. 3.3 µM or 10 µM) using two independent probes. The average fold of induction ± standard deviation is shown. N = 3 biological replicates with two technical replicates each. Comparisons were performed between treatments of every genotype and the corresponding treatment of wild‐type (WT) cells, ANOVA test (****P < 0.0001; ***P < 0.001; n.s. = non‐significant).
- Immunoblot analysis of samples treated as in (C). N = 3 independent experiments.
- Proposed model for the centrosome‐dependent PIDDosome activation upon different stimuli. The centrosome constitutively acts as PIDD1 centralizer. A local increase in PIDD1 concentration (achieved either by centrosome clustering or upon p53 activation) triggers PIDDosome activation. In ANKRD26‐deficient cells, the inability of the centrosome to generate a local increase in PIDD1 concentration hinders the activation of the complex in response to both stimuli.
Source data are available online for this figure.
