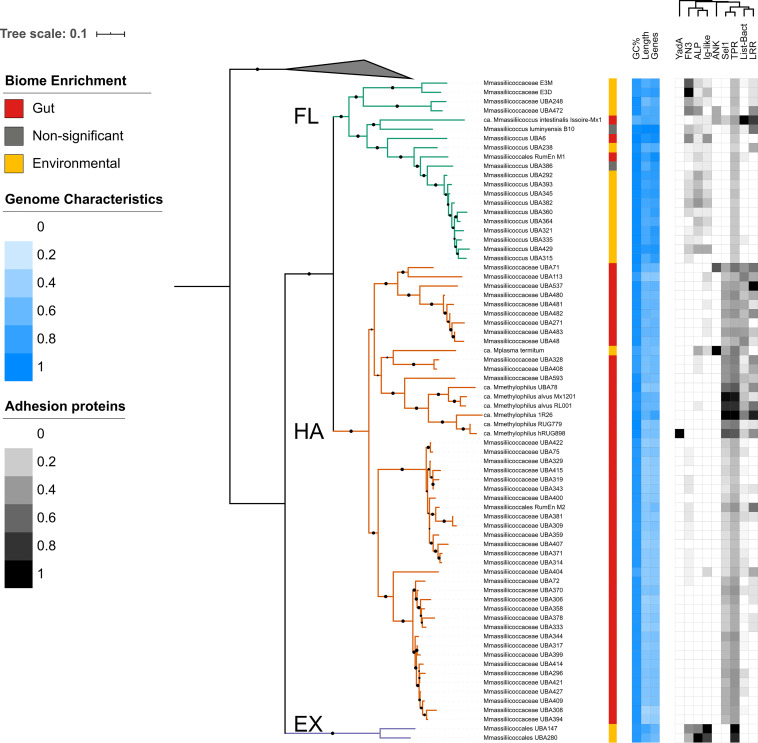
FIG 3.
Genome characteristics and adhesion proteins of Methanomassiliicoccales reflect division of the order into clades. Note that members of clade FL not enriched in environmental biomes resemble those of clade HA. The phylogeny is the same as that shown in Fig. 1. The colored strip summarizes the biome enrichment analysis. Heatmaps show genome features, including genome GC content (“GC%,” range, 41.26% to 62.74%), genome length (“Length,” 969.311 bp to 2.620.233 bp), and number of predicted genes (“Genes,” 1,057 to 2,607) (blue scale), or a repertoire of eukaryote-like proteins: YadA-like domain (YadA; 0, 1), fibronectin type III (FN3; 0, 20) domains, bacterial Ig-like domains (Ig-like; 0, 12), ankyrin repeats (ANK; 0, 3), Sel1-containing proteins (Sel1; 0, 29), tetratricopeptide repeats (TPR; 7, 40), Listeria-Bacteroides repeat-containing proteins (List-Bact; 0, 26), and leucine-rich repeats (LRR; 0, 9) (gray scale shows columns ordered by hierarchical clustering); and adhesion-like proteins (ALP; 0, 12). On both heatmaps, the color intensity of each feature is relative to the maximum value of each category.