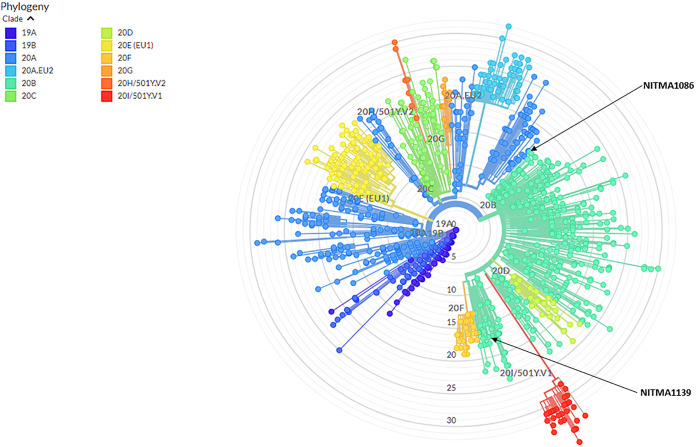
FIG 1.
Phylogenetic tree displaying sequenced SARS-CoV-2 strains (with reference to GISAID global data set), in accordance to the clades assigned for the virus during phylogenetic analysis using the Augur toolkit (with default parameters) run through the Nextstrain server. Augur implements the FastTree algorithm, which infers the maximum-likelihood method for building phylogenetic trees from alignments of nucleotide or protein sequences (7, 10). Both of the sequenced strains are colored according to the assigned clade, complying with the recently established nomenclature (11). The locations of the two isolates, NITMA1089 and NITMA1139, are marked with black arrows.