FIGURE 1.
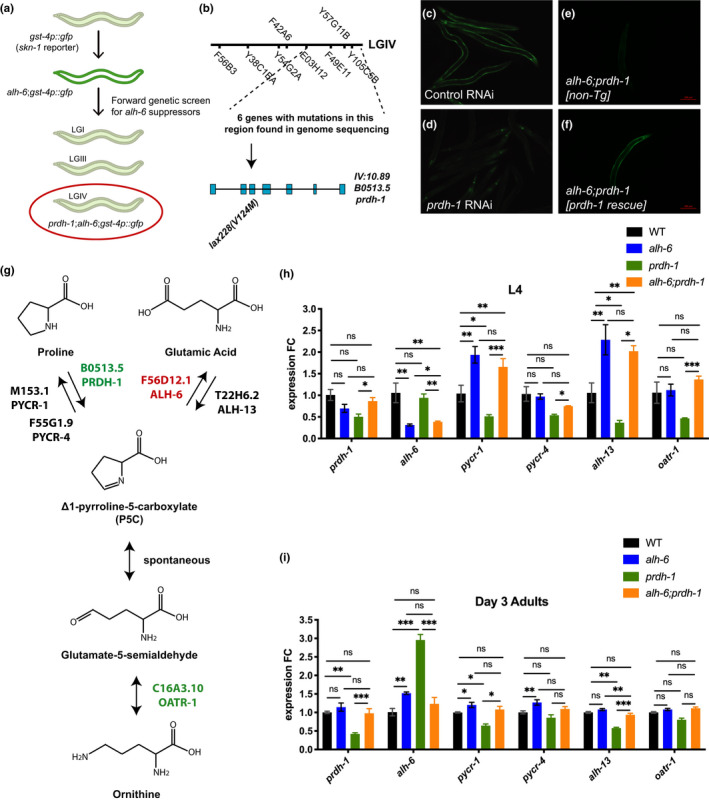
prdh‐1 mutation suppresses activation of SKN‐1 and proline metabolism deregulation in older alh‐6 animals. (a) Cartoon depiction of EMS screen for suppressors of SKN‐1 reporter activation in alh‐6 mutants. (b) SNP mapping identifies linked loci of prdh‐1(lax228) marked by dashed lines. Mutation locus of prdh‐1(lax228) in the gene is marked by arrow. (c, d) RNAi knockdown of prdh‐1 suppresses the activation of SKN‐1 in alh‐6;gst‐4p::gfp animals. (c) Day 3 adult alh‐6;gst‐4p::gfp fed L4440 (control RNAi). (d) Day 3 adult alh‐6;gst‐4p::gfp fed prdh‐1 RNAi. (e, f) prdh‐1 rescue reverts suppression of SKN‐1 activation in alh‐6(lax105);prdh‐1(lax228);gst‐4p::gfp animals. (e) Day 3 alh‐6(lax105); prdh‐1(lax228);gst‐4p::gfp adults. (f) Day 3 alh‐6(lax105);prdh‐1(lax228);gst‐4p::gfp adults with prdh‐1 rescue construct. (g) Schematic of biosynthetic and catabolic pathways of proline in C. elegans. (h,i) RT‐PCR analysis of gene expression changes in the proline metabolism pathway in L4 and Day 3 adult hermaphrodites. (h) alh‐6 single and alh‐6;prdh‐1 double mutant animals show similar increased expression in proline biosynthesis genes at L4 stage. (i) Day 3 alh‐6 mutant animal upregulate genes such as alh‐6, pycr‐1, and pycr‐4 in concerted effort to detoxify P5C. Notably, older alh‐6;prdh‐1 double mutants show WT level of expression in proline metabolism genes. Statistical comparisons of RT‐PCR results in worms were done using ANOVA between all groups. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. All studies performed in minimum of biological three triplicates; refer to Table S1 for n for each comparison
