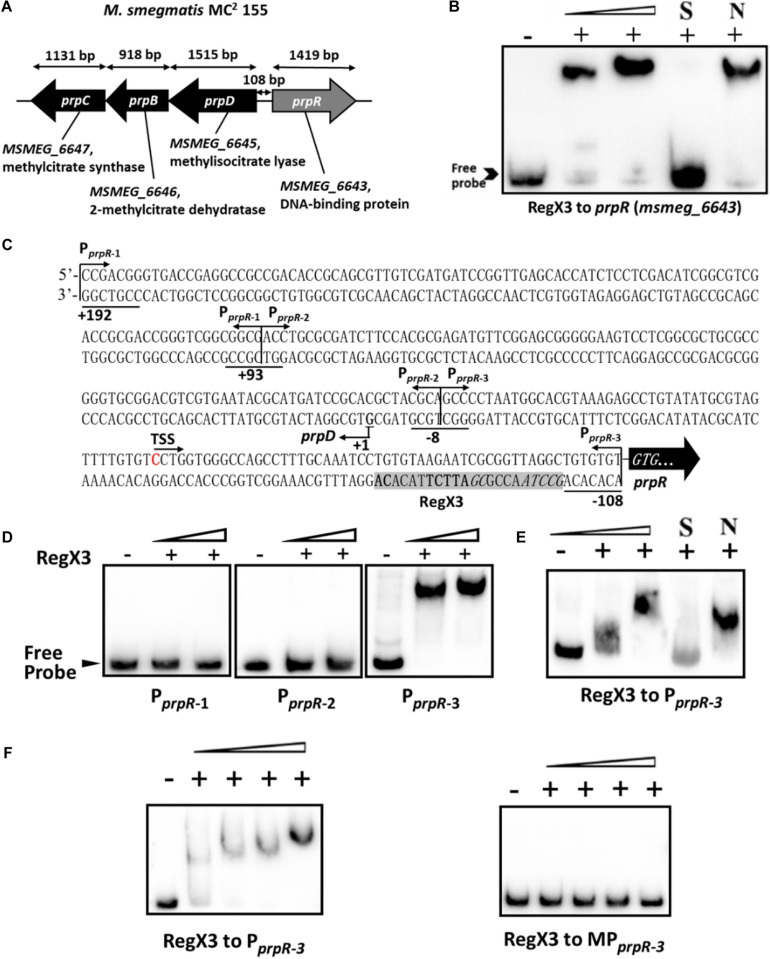
FIGURE 2.
Binding of RegX3 to the regulatory region of the genes related to the methylcitrate cycle. (A) The schematic depiction of prpDBC (msmeg_6645-6647) and prpR (msmeg_6643) regions on the M. smegmatis MC2 155 chromosome. The open reading frame prpR is located in close proximity to the prpDBC operon. (B) EMSA showing the binding of His-RegX3 with the promoter of prpR. The biotin-labeled probe, a 300-bp DNA fragment corresponding to the upstream region (-300 to 0 bp) of prpR, was co-incubated with different concentrations of recombinant His-RegX3 protein (0, 0.6, and 1.2 μM). S represents the unlabeled specific probe, N represents the non-specific competitor DNA (Salmon sperm DNA). (C) Putative binding site (shadowed) of RegX3 in the promoter regions of prpR. The +192, +93, -8, and -108 positions are underlined; the three short fragments (PprpR–1, PprpR–2, and PprpR–3) and TSS (transcriptional start site) are marked. (D) EMSA showing the binding of His-RegX3 (0, 3.6, and 7.1 μM) with three biotin-labeled short fragments from prpR upstream promoter region. (E) Binding of biotin labeled PprpR–3 with increasing concentration of His-RegX3 (0, 1.6, and 3.2 μM). (F) EMSA showing the binding of His-RegX3 (0, 0.35, 0.7, 1.4, and 2.8 μM) with biotin-labeled PprpR–3 containing predicted consensus binding motif (GCCTAACCGCGATTCTTACACA) (left) or biotin-labeled MPprpR–3 containing mutate binding motif (TCCTCACCGCGAGGATTACACA) (right).