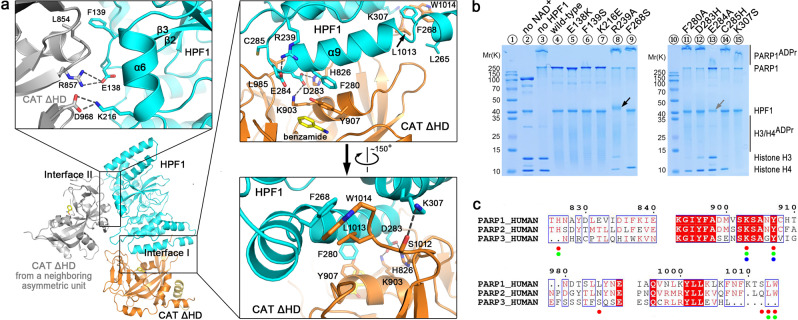
Fig. 2. Crystal structure of human HPF1/PARP1-CAT ΔHD and the binding mode between these two proteins.
a Overall structure and the interaction details between human HPF1 and PARP1-CAT ΔHD. The overall structure of human HPF1/PARP1-CAT ΔHD is shown as cartoons. HPF1 and PARP1-CAT ΔHD are colored in cyan and orange, respectively. The gray-colored molecule on the left side indicates another PARP1-CAT ΔHD molecule from a neighboring crystallographic asymmetric unit that also contacts the HPF1 molecule, shown here due to close packing of molecules in the crystal. Therefore, the complex crystal structure indicated two possible interaction modes between HPF1 and PARP1-CAT ΔHD. The three insets show the key residues on the two interfaces that mediate the interactions between the HPF1 and PARP1 in the crystal. The dashed lines indicate polar interactions between the key residues. b Mutagenesis/ADP ribosylation studies to verify which key residues indicated in the crystal structure mediate HPF1 and PARP1 binding in solution. Hyper-automodification of PARP1 (shown by the smeared PARP1ADPr bands on SDS-PAGE) was thought to indicate a loss of the interaction between HPF1 mutants and PARP1. Surprisingly, we noted that HPF1 R239A, but not any other HPF1 mutants, was robustly ADP-ribosylated in the assay (black arrow, also see Fig. 4c), while E284A seemed to be mildly ADP-ribosylated. The HPF1 E284A band became weaker, indicating a loss of unmodified protein in the assay (gray arrow). c Amino acid sequence alignment of human PARP1/2/3. The verified key residues that mediate HPF1 binding in PARP1 in solution are indicated by red dots. The corresponding residues in PARP2 and PARP3, if conserved in PARP1, are indicated by green and blue dots, respectively.