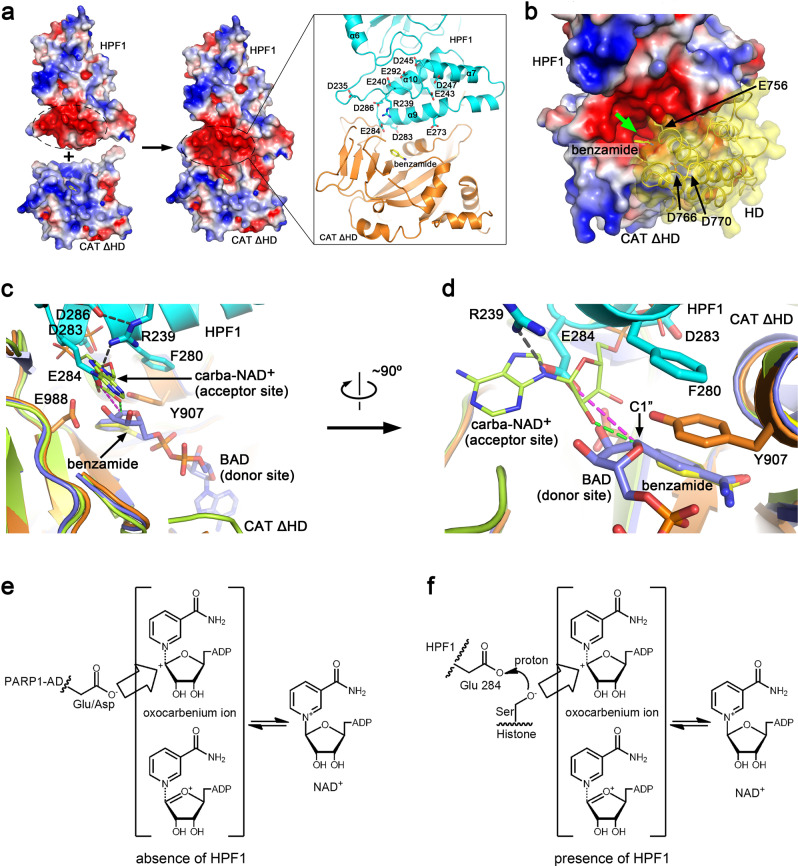
Fig. 3. Structural basis of the mechanism through which HPF1 modulates PAPR1 activity.
a Surface electrostatic potential of human HPF1, PAPR1-CAT ΔHD and the hetero-dimer. The negatively and positively charged regions are colored red and blue, respectively. A strongly negatively charged region in HPF1 merges with the active site of PARP1-ART in the complex, creating a joint negatively charged active site. The inset on the right shows the acidic and basic residues in this negatively charged region of HPF1. b A close view of the remodeled active site and the HD domain if in the folded state. The HD domain is shown as cartoons covered by transparent surface based on a superimposition of the ART domain from the HPF1/PAPR1-CAT ΔHD structure and the ART domain from the DNA-bound PARP1 crystal structure (PDB 4DQY). The green arrow indicates the entrance to the tunnel leading to the remodeled active center. c, d Superimposition of our HPF1/PAPR1-CAT ΔHD complex structure (cyan and orange) with the previously reported PARP1-ART/BAD complex structure (PDB 6BHV, light-blue)22 and the PARP1-CAT/carba-NAD+ complex structure (PDB 1A26, lime)29. Two different views of the active cite are shown. BAD (light-blue sticks) is an NAD+ analog that mimics the ADP-ribosylation donor. The ADP moiety of carba-NAD+, shown as lime sticks in this figure, has been proposed to represent the ADP-ribosylation acceptor for ADPr chain elongation/branching. e A proposed catalytic mechanism of PARP1 automodification in the absence of HPF1. Without HPF1, the acidic residues in automodification domain (AD) of PARP1 can access the active center to get ADP-ribosylated. f Proposed catalytic mechanism of histone serine ADP-ribosylation in the presence of HPF1. Upon HPF1 binding, HPF1 Glu284 carboxyl is positioned approximately 4.6 Å (see the purple dashed lines in c and d) away from the donor NAD+ ribose (represented by BAD). This distance is too great for Glu284 itself to accept the ADP-ribose, but could catalyze ADP-ribosylation of a serine by promoting deprotonation of its sidechain hydroxyl.