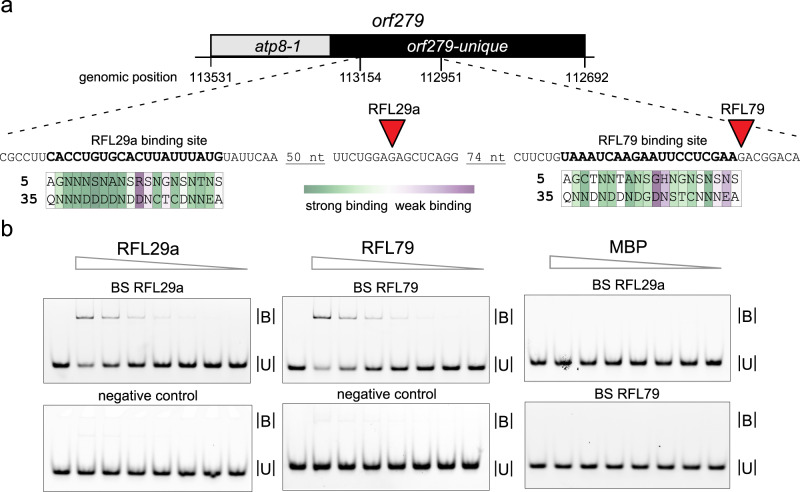
Fig. 7. RNA-binding assays with recombinant RFL29a and RFL79 proteins.
a Location of the predicted RFL29a and RFL79 binding sites within the orf279 RNA, relative to the observed cleavage sites (red triangles). The predicted binding site for RFL79 encompasses nucleotides 112,975–112,956, and for RFL29a 113,148–113,129 in the T. timopheevii mitochondrial genome (GenBank accession: NC_022714). The key amino acids at positions 5 and 35 of each PPR motif are indicated. The colour scale reflects the strength of the match between the amino acid combination and the RNA base, calculated from in vitro binding data43. b RFL29a and RFL79 bind to orf279 as shown by electrophoretic mobility shifts of fluorescein-labelled RNA oligonucleotides corresponding to their predicted targets. B = bound (RNA + protein) U = unbound (free RNA probe). Neither protein binds to an unrelated RNA oligonucleotide (based on orf256 sequence). As both proteins were produced as maltose-binding protein (MBP) fusions, RNA binding with MBP alone was tested and found to be negligible. Serial protein dilutions ranging from 1.8 µM to 28.1 nM for RFL29a and RFL79, and from 0.5 µM to 7.8 nM for MBP were used for the binding assays. The final concentration of the RNA probes was 1 nM. On each gel, the left lane acts as a marker for unbound probe. More details are provided in Supplementary Fig. 7. Probe sequences are given in Supplementary Table 6. This experiment was performed three times with similar results. Source data underlying Fig. 7b are provided as a Source data file.