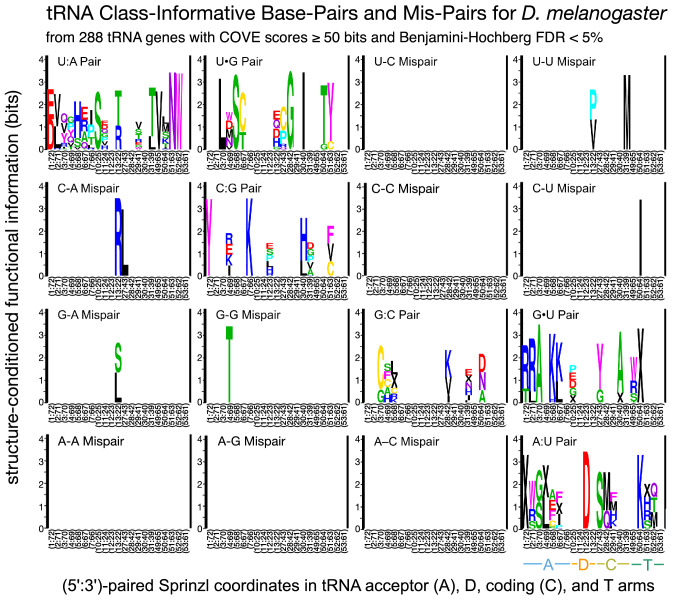
Fig. 4.
Class-Informative Base-Pairs and Base Mis-Pairs from 288 re-annotated tRNA genes in Drosophila melanogaster with COVE scores of at least 50 bits as computed in tSFM v1.0.1 with a Benjamini–Hochberg False Discovery Rate of 5% (CIFs not meeting this significance threshold are not shown). The total height of a stack of letters quantifies information gained about the functional type of a tRNA by a tRNA-binding protein if it specifically recognizes the paired features indicated. The letters within each stack symbolize functional types of tRNA genes, wherein IUPAC one-letter amino acid codes represent elongator tRNA aminoacylation identities and “X” symbolizes initiator tRNAs. The relative heights of letters within each stack quantify the over-representation of tRNA functional types carrying that feature relative to the background frequency determined by the frequencies of genes of various functional types (as calculated through normalized log-odds)