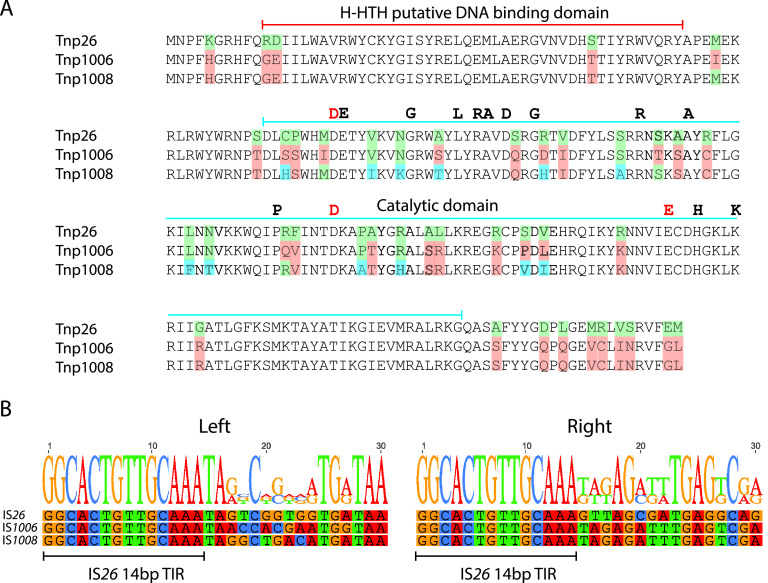
FIG 1.
Comparison of IS26, IS1006, and IS1008. (A) Alignment of the amino acid sequences of the transposases of IS26, IS1006, and IS1008. Green, blue, and orange indicate amino acids that differ in one or more of the sequences. The extents of the H-HTH putative DNA binding domain and the DDE catalytic domain are marked above the sequences. The completely conserved DDE residues are marked by red letters, and residues conserved in >90% of the broader IS26 family (1) are marked by black letters. The K180 residue, which is an R in one clade of the IS26 family (1), is also marked. (B) Alignment of 30 bp of sequence at the left and right ends of IS26, IS1006, and IS1008. The sequences of the right ends have been reversed and complemented for ease of comparison. The 14-bp terminal inverted repeats (TIR) are marked by bars.