Abstract
The introduction of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into the human population represents a tremendous medical and economic crisis. Innate immunity—as the first line of defense of our immune system—plays a central role in combating this novel virus. Here, we provide a conceptual framework for the interaction of the human innate immune system with SARS-CoV-2 to link the clinical observations with experimental findings that have been made during the first year of the pandemic. We review evidence that variability in innate immune system components among humans is a main contributor to the heterogeneous disease courses observed for coronavirus disease 2019 (COVID-19), the disease spectrum induced by SARS-CoV-2. A better understanding of the pathophysiological mechanisms observed for cells and soluble mediators involved in innate immunity is a prerequisite for the development of diagnostic markers and therapeutic strategies targeting COVID-19. However, this will also require additional studies addressing causality of events, which so far are lagging behind.
Keywords: COVID-19, SARS-CoV-2, pandemic, innate immunity, interferon, viral sepsis, immunosuppressive cells, monocytes, granulocytes, genetics, trained immunity
Differences in innate immune system function may underpin the curiously variable disease spectrum of COVID-19.
Introduction
Coronavirus disease 2019 (COVID-19) (Berlin et al., 2020; Gandhi et al., 2020), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Kim et al., 2020; Wu et al., 2020; Yao et al., 2020; Zhang and Holmes, 2020) is a rather heterogeneous disease. Disease courses range from mainly asymptomatic and mild courses to more severe and critical courses in 10%–20% of symptomatic patients who are at considerable risk of fatality with many different organ systems involved in differing combinations and with variable symptoms (Gupta et al., 2020) (Box 1 ). The virus itself, the environment, and the host can contribute to such disease heterogeneity (Morens and Fauci, 2020). Numerous mutations identified so far by thousands of viral sequences obtained during the first 9 months of the pandemic (Candido et al., 2020; Oude Munnink et al., 2020) suggest that viral genetic diversity, genetic evolution, variable infectivity, or co-pathogenesis might contribute to infectivity and fatality, but not so much to disease heterogeneity observed for COVID-19. Although many mutations decreased infectivity, the virus variant D614G in the spike protein of SARS-CoV-2 increased infectivity (Li et al., 2020). Two clades, recently emerged in the United Kingdom (B.1.1.7) and South Africa (B.1.1.351), similarly increased infectivity. Overall, however, there is little evidence for the virus to be mainly responsible for disease heterogeneity, which leaves the host and the environment as factors influencing disease course and outcome. Although there is evidence that environmental factors such as seasonality, other natural disasters, environmental degradation, quality of the public health infrastructure, or governance by state authorities have an impact on overall disease burden for society (Han et al., 2020), the host itself seems to be the major factor explaining disease severity, infection rates (Merad and Martin, 2020; Vabret et al., 2020), and long-term medical consequences (Marshall, 2020). Disease severity and mortality rates are significantly higher in elderly populations pointing toward lack of widespread long-lasting pre-existing adaptive immunity by T or B cells against SARS-CoV-2 (Morens and Fauci, 2020). This is in stark contrast to the 2009 influenza pandemic, which had much lower incidence rates in the elderly population, indicating partial protection on population level due to earlier exposure to similar influenza viruses (Xu et al., 2010). It is possible that cross-protection via adaptive immune responses against endemic coronaviruses might be in part responsible for mild disease courses in younger individuals (Lipsitch et al., 2020). An effective immune response against SARS-CoV-2 requires both arms of the immune system, the innate immune system (Box 1) including granulocytes, monocytes, and macrophages among other cells of the innate immune system and the adaptive immune system with T and B cells (see Sette and Crotty [2021] for adaptive immunity against SARS-CoV-2). A role of the innate immune system in determining disease severity and outcome is supported by recent genetic findings (van der Made et al., 2020; Severe Covid-19 GWAS Group et al., 2020; Zhang et al., 2020a), interaction maps of viral proteins with host factors (Gordon et al., 2020), or high resolution single-cell omics analyses (Chua et al., 2020; Schulte-Schrepping et al., 2020). Here, we provide a conceptual framework for determining host-virus interactions of a pandemic threat with an emphasis on the innate immune system, highlight important findings about the innate immune system in COVID-19, and suggest avenues for further research on the most prioritized open questions.
Box 1. Definition for disease severity, the innate immune system, and systems medicine.
COVID-19 disease severity categories. In addition to national categories the WHO disease severity category is widely used. Here, the major categories are mild disease, moderate disease (mainly characterized by pneumonia), severe disease (with severe pneumonia), and critical disease (with acute respiratory distress syndrome [ARDS] and/or sepsis and/or septic shock). A number of other complications have been described, particularly for severe and critical disease such as acute pulmonary embolism, acute coronary syndrome, acute stroke or delirium among many others including Guillain-Barré syndrome.
Innate immune system, innate immune cells, and innate immunity. The immune system is divided into two arms, the adaptive immune system with T and B lymphocytes and the innate immune system with all other immune cell types of which granulocytes, monocytes, macrophages, and NK cells are the more common cells, but many other innate immune cells also exist including different dendritic cells, innate lymphoid cells, or mast cells. Although the majority of all species possess an innate immune system, the conventional adaptive immune system is restricted to jawed vertebrates. Whereas most immune responses involve both the innate immune system and the adaptive immune system evoking innate and adaptive immunity, respectively, the extent to which both arms are recruited to an immune response differs between stimuli.
Systems Medicine. In this review, we use the term Systems Medicine as the implementation of systems biology approaches in medical research and practice. A given hypothesis concerning e.g., the role of an organ system such as the immune system in a disease such as COVID-19 is answered by an iterative circular process between (I) clinical investigations, (II) computational multiscale modeling, followed by (III) experimental validation of (1) pathogenic mechanisms, (2) disease progression versus remission, (3) cure or disease spread, and (4) treatment responses and adverse events, both at the population and the individual patient level. High-resolution, high-throughput, and high-content technologies, particularly the omics technologies, are often used to reach the goals of Systems Medicine. The long-term goal of Systems Medicine is to provide measurable improvements for the patients’ health.
Conceptualizing the interaction between the host immune response and the virus
Looking at the current pandemic entirely from a biological perspective and assuming that the majority of humankind does not have an extended cross-reaction to other viruses of the coronavirus family (Braun et al., 2020; Grifoni et al., 2020; Mateus et al., 2020), the introduction of SARS-CoV-2 into the human population is one of the biggest evolutionary events in the last hundred years (Morens and Fauci, 2020; Morens et al., 2020). In such an evolutionary experiment, the innate immune system must be assigned a very special role in the defense against SARS-CoV-2 (Amor et al., 2020; Mantovani and Netea, 2020; Vabret et al., 2020). An important part of innate immunity is the cell-autonomous response of the cells that get infected by the virus, which is influenced by the biology of the receptors and co-receptors for viral entry (Hoffmann et al., 2020; Wu et al., 2020; Yao et al., 2020), as well as all cellular mechanisms that determine the viral life cycle (Cyranoski, 2020). In the SARS-CoV-2 pandemic, the virus encounters the human population as a swarm of genetic variants and the processes of infection and virus replication may still be subject to major genetic changes, by which certain virus variants may achieve an evolutionary advantage. These processes are in full swing and accelerate as more individuals become infected (Callaway, 2020; Cyranoski, 2020; Korber et al., 2020). It therefore will be important to monitor infected individuals for potential changes in the very early steps of the immune response to the virus, mainly triggered by infected cells and early interactions with adjacent innate immune cells.
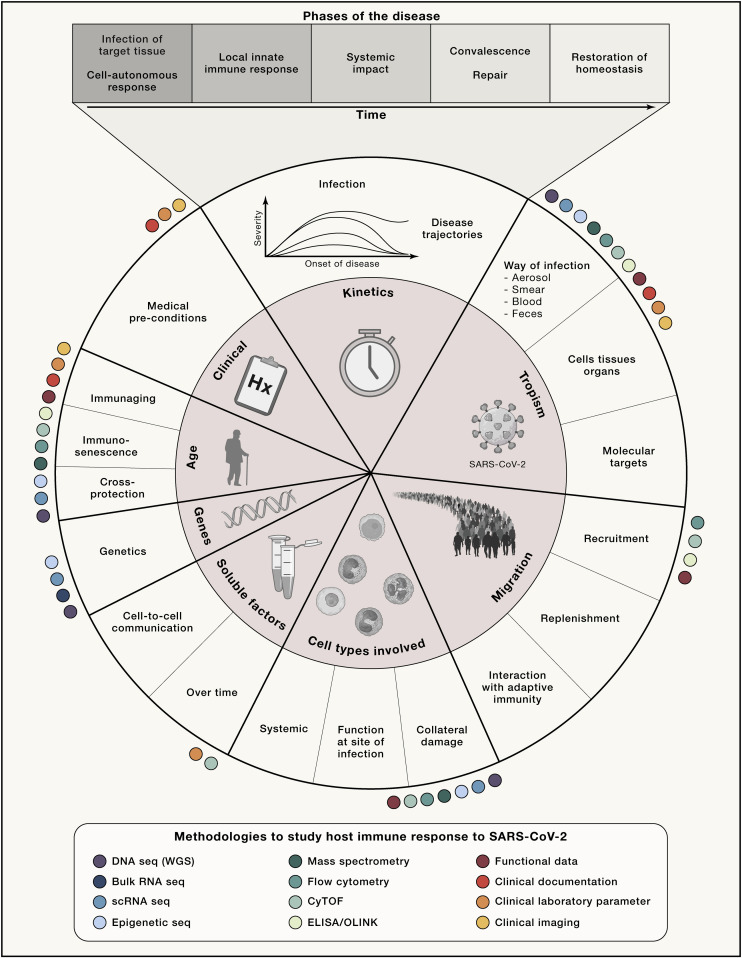
Conceptually, when addressing important defense mechanisms of the host, it is important to consider all parameters that may be relevant for such an initial interaction of a new virus with a species (Figure 1 ). Bearing in mind that most of the steps of the interaction between the virus and the host will follow a normal distribution of attributes or parameters required to describe the outcome of the interaction, it is not entirely surprising that the clinically observed disease courses show an enormous heterogeneity (Berlin et al., 2020; Gandhi et al., 2020). For example, the induction of an interferon response by an infected cell might follow a normal distribution in the population with low, intermediate, and high responders thereby triggering different magnitudes of cellular responses and thus very different downstream effects (e.g., in the innate immune system). In Figure 1, we illustrate important innate immune mechanisms that might be particularly prone to heterogeneous outcomes—due to both environmental and genetic factors—within the human population and therefore these should be a major focus in our efforts to determine the role of the innate immune system for infectivity, viral spreading, and disease course but also long-term outcome. We postulate that such conceptualization of the interplay between the innate immune system and the virus will help to focus on the most critical steps first, which then can be validated in larger studies.
Figure 1.
Conceptualizing the interaction between the host immune response and the virus
Proposed fields of research along the disease trajectory in five phases that influence pathophysiology with an emphasis on innate immunity. Methodologies suggested to be applied for addressing certain areas are represented as color-coded circles. These reflect frequently used methods in previously published studies on COVID-19. This is only a selection and we make no claim to completeness. The overall concept could be extended to the adaptive immune system and other organ systems. CyTOF, cytometry by time of flight, mass cytometry; ELISA; OLINK, plasma proteome by proximity extension assay; scRNA-seq, single-cell RNA sequencing; seq, sequencing; WGS, whole genome sequencing.
As we will discuss, hypothesis-driven systems medicine approaches (Box 1) have a very high chance to quickly uncover the most critical and variable steps in the interactions between the virus and the host’s immune system to link them to the different clinical phenotypes allowing a better stratification of patients that will foster the derivation of therapeutic procedures (Rajewsky et al., 2020).
The breach—point of entry for SARS-CoV-2
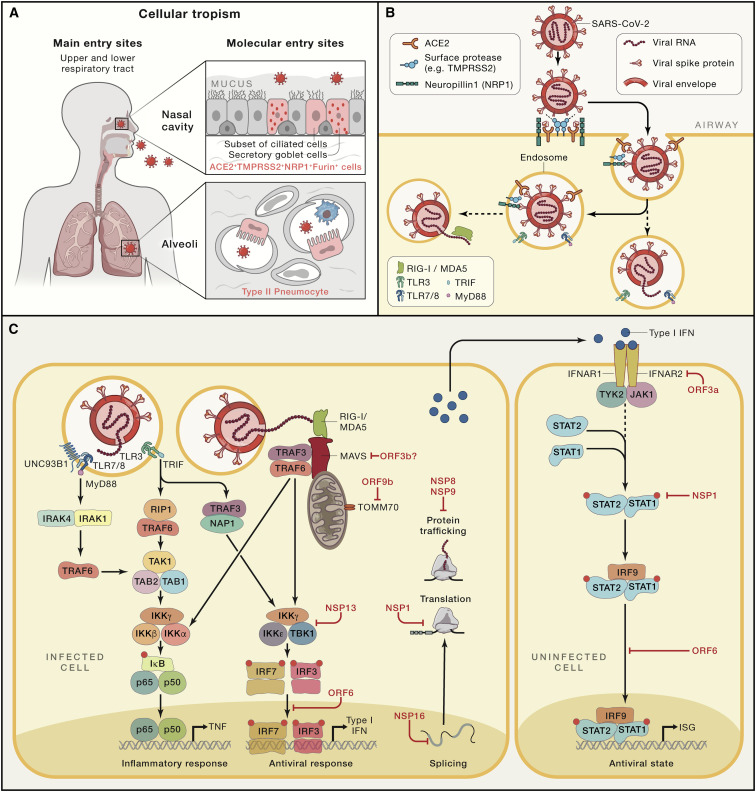
Major determinants of the host’s innate immune response are dictated by the cell tropism of the virus and its ability to circumvent innate immune responses (Morens and Fauci, 2020). With the identification of SARS-CoV-2 (Hoffmann et al., 2020; Wu et al., 2020; Yao et al., 2020), and based on its close relationship with the SARS coronavirus (SARS-CoV) (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020), it became quickly clear that the surface receptor for SARS-CoV (Li et al., 2003), the angiotensin-converting enzyme 2 (ACE2), was also a major cellular entry point for SARS-CoV-2 (Hoffmann et al., 2020). Similar to SARS-CoV (Matsuyama et al., 2010), SARS-CoV-2 employs the cellular serine protease TMPRSS2 for S protein priming. Most recently, Neuropilin1 (NRP1) was identified as an important cofactor for entry, particularly in cells with low level ACE2 expression (Cantuti-Castelvetri et al., 2020). Several other proteases including Furin have also been indicated as co-factors (Ou et al., 2020).
In absence of cell-type-specific expression maps of SARS-CoV-2 viral entry-associated genes, the Human Cell Atlas (HCA) Lung Biological Network investigated the prevalence of ACE2 and TMPRSS2 in the body by analyzing their expression in single-cell RNA sequencing data (scRNA-seq) from multiple tissues from healthy human donors (Sungnak et al., 2020). Probably most important for the high efficacy of SARS-CoV-2 transmission is the high co-expression of ACE2 and TMPRSS2 in nasal epithelial cells, particularly in two types of goblet cells and a subset of ciliated cells, showing the highest expression among all cells in the respiratory tree (Figure 2 A). An interesting observation of this study was the high co-expression of genes involved in early steps of antiviral immune responses in these epithelial cells, which indicated the potential for these specialized nasal cells to play an important role in the initial viral infection, spread, but potentially also, clearance. In the lung, ACE2 and TMPRSS2 expression has been mainly identified in alveolar epithelial type II cells (Qi et al., 2020; Zou et al., 2020). TMPRSS2 is more widely expressed than ACE2, and cells with simultaneous expression were identified in the respiratory tree, the cornea, esophagus, ileum, colon, gallbladder, and common bile duct (Sungnak et al., 2020), yet there was no evidence for simultaneous expression in immune cells. Extending these studies to even larger datasets, including dozens of different scRNA-seq datasets derived from many groups around the world, revealed the power of metadata analysis based on scRNA-seq (Muus et al., 2021). Here, it was even possible to predict the impact of clinical risk factors such as sex, age, and smoking history among others on gene expression level of genes associated with viral entry. A second study from the HCA Lung Biological Network extended the initial findings to tissues from non-human primates and mice, indicating co-expression of ACE2 and TMPRSS2 within nasal goblet secretory cells, lung type II pneumocytes, and ileal absorptive enterocytes (Ziegler et al., 2020). In contrast to earlier reports suggesting ACE2 to be an interferon-stimulated gene (ISG), more recent evidence showed that a truncated form of ACE2 designated deltaACE2, but not ACE2 itself, is an ISG (Onabajo et al., 2020). DeltaACE2 neither binds the SARS-CoV-2 spike protein nor serves as a carboxypeptidase.
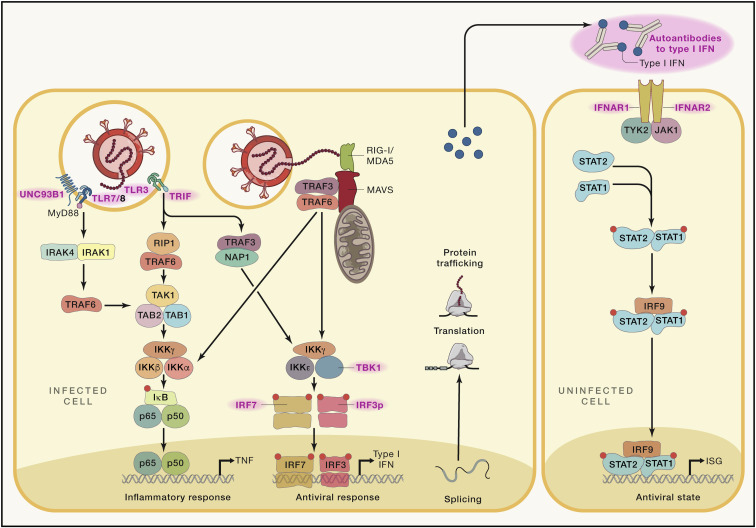
Figure 2.
SARS-CoV-2 tropism, infection, and alarming the innate immune system
(A) Major entry sites of SARS-CoV-2 via cells within the nasal cavity and the upper and lower respiratory tract.
(B) Molecular determinants during SARS-CoV-2 infection of a cell.
(C) SARS-CoV-2 is most likely recognized by PRRs recognizing foreign RNA including endosomal TLR3 and TLR7 as well as by cytoplasmic RIG-I and MDA5. Predicted downstream signaling events based on findings from genetic studies, functional and clinical observations, interaction mapping, or CRISPR screens. Interactions between SARS-CoV-2-derived proteins and cellular mechanisms or in case of interaction mapping derived information, detection of direct interaction between viral or host proteins. ORF3b was functionally determined to suppress type I IFN, but a direct target was not identified (Konno et al., 2020). ACE2, angiotensin-converting enzyme 2; IFNAR1, interferon-alpha/beta receptor alpha chain; IκB, inhibitor of κB; IKKα/ꞵ/γ/ε, IκB kinase α/ꞵ/γ/ε; IRAK1/4, interleukin-1 receptor-associated kinase 1/4; IRF3/7/9, interferon regulatory factor 3/7/9; ISG, interferon-stimulated genes; MDA5, melanoma differentiation-associated protein 5, RIG-I-like receptor dsRNA helicase enzyme; MyD88, myeloid differentiation primary response 88; NAP1, NF-κB-activating kinase-associated protein 1; NRP1, neuropilin 1; NSP, non-structural proteins of SARS-CoV-2; ORF, open reading frames of SARS-CoV-2; p50/65, the two subunits of NF-κB; RIG-1, retinoic acid-inducible gene I, a cytoplasmic pattern recognition receptor recognizing double-stranded RNA; RIP1, receptor interacting serine/threonine kinase 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; STAT1/2, signal transducer and activator of transcription 1/2; TAB1/2, TGF-beta activated kinase 1 binding protein 1/2; TAK1, TGF-beta activated kinase (encoded by MAP3K7); TBK1, TANK-binding kinase 1; TLR3, Toll-like receptor 3, CD283; TLR7/8, complex of Toll-like receptors 7 and 8, TLR7 also known as CD287 and CD288, respectively; TMPRSS2, transmembrane protease, serine 2; TRAF3/6, TNF receptor-associated factor 3/6; TRIF, TIR-domain-containing adaptor-inducing interferon-β; TYK2, tyrosine kinase 2; UNC93B1, Unc-93 homolog B1.
By applying a newly developed computational pipeline called Viral-Track, viral RNA was detected in single-cell transcriptomes of bronchoalveolar lavage fluid (BALF) samples from COVID-19 patients with severe, but not mild, disease course (Bost et al., 2020), suggestive of a differential progression of infection in the lung. The strongest enrichment of viral RNA was observed in ciliated and epithelial progenitors. Viral RNA was also detected in a subset of macrophages (SPP1 + macrophages) (Bost et al., 2020; Chua et al., 2020). Whether these myeloid cells are directly infected or whether these cells phagocytosed cellular material carrying viral RNA requires further investigation. In vitro monocyte-derived macrophages and dendritic cells (DCs) are susceptible to SARS-CoV-2 infection, which is abortive, because these cells do not support virus replication (Yang et al., 2020a). Interestingly, as it has also been suggested that potential co-infections might determine the clinical phenotype and disease outcome, co-infection of monocytes in the BALF with human metapneumovirus was identified in one of the severe COVID-19 patients (Bost et al., 2020), and it was postulated that such co-infections might explain—at least in part—critical COVID-19 disease courses. However, much larger studies are urgently needed to verify these important, but still preliminary, findings.
Information about the role of innate immune cells as potential targets of SARS-CoV-2 or mediating cells for the infection in organs or tissues other than the respiratory tract is still sparse. Within the oral cavity, there is little evidence for expression of ACE2, TMPRSS2, or Furin on any other cells than epithelial cells (Sakaguchi et al., 2020). Furthermore, there has been no evidence for the role of innate immune cells in SARS-CoV-2 gaining access to the retina (de Figueiredo et al., 2020) or the brain (Song et al., 2021), albeit the virus has been found in both organs. Similarly, the human placenta is highly susceptible to SARS-CoV-2 infection, but only non-immune cells including syncytiotrophoblasts in the first trimester and extravillous trophoblasts in the second trimester express ACE2 and TMPRSS2 and have been identified as cellular targets for the virus (Ashary et al., 2020). Interestingly, other coronavirus family member receptors (ANPEP and DPP4) and several proteases (CTSL, CTSB, and Furin) are uniformly expressed within the placenta throughout the first and second trimester, and genes necessary for viral budding and replication were also identified, strongly suggesting that the placenta is a vulnerable target for productive infection (Ashary et al., 2020). Yet, based on clinical observations, there is no evidence for vertical transmission of the virus from infected mothers to the fetus (Islam et al., 2020) arguing for a protective role of the maternal immune response, which requires further exploitation.
An interesting finding is the expression of ACE2 and TMPRSS2 on platelets, considering that patients present with signs of platelet hyperactivation, particularly in severe COVID-19 (Zhang et al., 2020b). In vitro exposure to SARS-CoV-2 or its Spike protein potentiated platelet aggregation, release of dense granules, upregulation of activation markers (PAC-1, CD62P), platelet spreading, and clot retraction. Furthermore, the virus facilitates the release of coagulation factors, the secretion of inflammatory cytokines, and the formation of leukocyte-platelet aggregates, which might be important mechanisms leading to thromboembolic complications observed in severe COVID-19 (Ackermann et al., 2020; Gupta et al., 2020; Zhang et al., 2020b). Whether other receptors including CD147 (BSG) can facilitate viral entry remains to be seen. CD147 is widely expressed and receptor expression and viral load does not seem to correlate (Bost et al., 2020) arguing against CD147 as a major entry receptor for SARS-CoV-2.
Collectively, there is still little evidence that innate immune cells are a primary target of productive infection of SARS-CoV-2. The detection of viral RNA in myeloid cells derived from lung or BALF of severe COVID-19 patients still allows several explanations, including phagocytosis of cellular material derived from infected cells, which requires further assessment. Similarly unclear is whether platelets in the blood might serve as a sponge for viruses, which could explain why viruses are rarely detected in blood specimens, yet seem to infect many organs.
Cytokine alterations and innate immune cell responses following infection
Alarming the immune system—recognition, interferon response, and immune evasion
A common pattern of cell entry via endocytosis following receptor binding was established for coronaviruses, which is followed by viral RNA release into the cytoplasm, production of viral proteins, and formation of viral replication/transcription complexes on double-membrane vesicles (Snijder et al., 2020). Although viral entry mechanisms are well established for SARS-CoV-2 (Shang et al., 2020), the following steps are not yet entirely resolved. Similarly, the pattern recognition receptors (PRRs) involved in recognizing SARS-CoV-2 are not yet directly determined. Based on findings from other coronaviruses, the most likely candidates are Toll-like receptors 3 (TLR3) and TLR7 in the endosome (Mazaleuskaya et al., 2012) or the cytosolic sensors retinoic acid-inducible gene 1 (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) (Sa Ribero et al., 2020) recognizing foreign viruses’ RNA (Figure 2B). All these PRRs are linked via signaling cascades to induce strong interferon responses. RIG-I and MDA5 trigger the downstream adaptor mitochondrial antiviral signaling protein (MAVS) on mitochondria followed by activation of tumor necrosis factor receptor-associated factor 3 (TRAF3), TRAF family member-associated nuclear factor κB (NF-κB) activator binding kinase 1 (TBK1), together with inhibitor of NF-κB kinase-ε (IKK), which leads to phosphorylation of the “master regulators” IRF3 and IRF7. Subsequently, transcription of type I IFNs and numerous ISGs are induced (Loo and Gale, 2011). Similar signaling cascades are known downstream of TLR3 and TLR7 (Figure 2C). Secreted type I IFNs signal via interferon receptors (IFNAR) to switch on Janus kinase 1 (JAK1) and tyrosine kinase 2 (Tyk2), activating STAT1, STAT2, and IRF9, which form a complex and translocate to the nucleus to bind to interferon-stimulated response elements (ISREs) in the genome. Hundreds of ISGs with various antiviral functions get induced, which can lead to further amplification loops by ISGs. Importantly, the type I IFN response can be rather heterogeneous. Depending on the state of the reacting cell (e.g., with respect to its metabolic or general activation state) (Talemi and Höfer, 2018), it is at least in part stochastic (Rand et al., 2012; Wimmers et al., 2018), it varies among different cell types and microenvironments, and it shows interindividual heterogeneity concerning the amplitude and kinetics (Patil et al., 2015). The ability to fine-tune an IFN response is critical because both its overactivation and underactivation are deleterious to the host (Mesev et al., 2019). The role of these fine-tuning mechanisms has not yet been addressed in detail in context of COVID-19.
Keeping the complex regulation of the IFN system in mind, it is critical to understand the dynamics of the type I IFN response in COVID-19. In in vitro infection models utilizing cell lines, SARS-CoV-2 induced only low type I and II IFNs, consequently inducing only moderate levels of ISGs and a unique proinflammatory cytokine signature including IL1B, IL6, and TNF, as well as many chemokines (CCL20, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL16) (Blanco-Melo et al., 2020). An early study in patients reported that type I IFNs were either not detected (particularly IFNβ) irrespective of disease severity or at lower levels (IFNα) in plasma mainly derived from patients with severe disease (Hadjadj et al., 2020). These observations were further extended by illustrating a lack of expression of genes encoding type I IFNs in peripheral blood mononuclear cells (PBMCs) of COVID-19 patients and an early but transient wave of ISG expression in blood-derived immune cells, which correlated with an early burst of IFNα, likely of lung origin (Arunachalam et al., 2020). An early transient wave of ISG expression in cells of the innate immune system, particularly monocytes, was also determined by scRNA-seq (Schulte-Schrepping et al., 2020). A longitudinal analysis confirmed an early peak with subsequent decline of IFNα and IFNλ in mild-to-moderate COVID-19, whereas levels further increased particularly during the second week in severe patients (Lucas et al., 2020). The dynamics of type I IFNs in severe patients is in line with findings in a murine model of SARS-CoV-2 infection illustrating that type I IFNs do not control SARS-CoV-2 replication in vivo but are significant drivers of pathologic responses (Israelow et al., 2020).
As an immune evasion strategy, SARS-CoV-2 uses a multipronged strategy to antagonize the IFN system with the clinical consequence of an insufficient type I IFN response having been recognized in early COVID-19 studies (Acharya et al., 2020; Blanco-Melo et al., 2020; Sa Ribero et al., 2020). At least ten SARS-CoV-2 proteins have been identified that counteract the antiviral action of IFN (Figure 2C). SARS-CoV-2-derived non-structural protein 16 (NSP16) suppresses global mRNA splicing and diminishes recognition of viral RNA by intracellular helicase receptors, NSP1 leads to global inhibition of mRNA translation by binding to the 18S ribosomal RNA in the mRNA entry channel, and NSP8 and NSP9 interfere with protein trafficking to the cell membrane; all three mechanisms independently leading to reduction of type I IFN production by the affected cell (Banerjee et al., 2020; Gordon et al., 2020). Interaction mapping further suggested that NSP13 interacts with TBK1, NSP15 with NRDP1 (an E3 ubiquitin ligase controlling the balance between JAK2-associated cytokine receptor degradation and ectodomain shedding), ORF9b with TOMM70 (an important receptor of the outer mitochondrial membrane), and ORF6 with either KPNA2 or the IFN-induced NUP98-RAE1 nuclear export complex, which are both involved in nuclear translocation of proinflammatory transcription factors such as IRF3, IRF7, or STAT1, a mechanism targeted by numerous other viruses including influenza, all antagonizing IFN signaling (Gordon et al., 2020; Xia et al., 2020). Functional testing further supported the counteracting activity of ORF3b, ORF6, NSP1, or NSP13 on type I IFN activation, of which particularly ORF6 suppressed STAT1 and STAT2 phosphorylation and STAT1 nuclear translocation, (Konno et al., 2020; Lei et al., 2020; Xia et al., 2020).
Accumulating evidence indicates that SARS-CoV-2 is targeting the type I IFN system at multiple steps thereby strongly interfering with a well-orchestrated interplay between antiviral and proinflammatory innate and adaptive defense mechanisms within the immune system. Studies with more in-depth longitudinal profiling and stratification by disease severity are warranted to clarify the questions surrounding the nature of the early IFN response to SARS-CoV-2. Further dissecting this delicate balance at the early steps of a SARS-CoV-2 infection for each patient individually might be critical to better stratify patients and to allow for patient-tailored treatment options thereby preventing the development of immunopathology as the basis for severe disease courses (Lee and Shin, 2020).
The next phase—from “cytokine storm” to immunosuppression
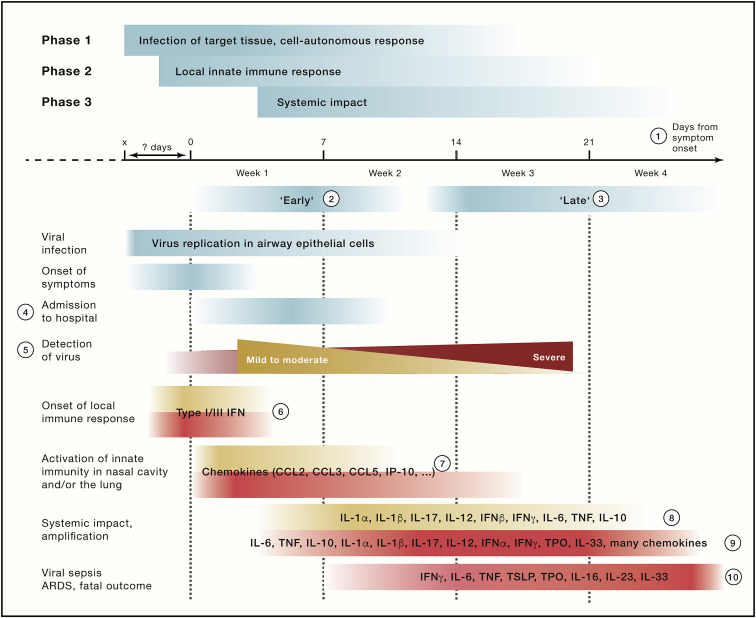
Conceptually, soluble factors including cytokines, chemokines, growth factors, inhibitory factors, hormones, and metabolites influence innate immune cells locally and systemically. Innate immune cells serve both as sender and receiver for soluble mediators. Soluble factors are important mediators shaping disease courses by triggering the release of innate immune cells from the bone marrow into circulation, their recruitment to inflamed and infected tissues, as well as by influencing their local motility and function. The combination, as well as the timing (kinetics) and the concentrations (dynamics) of these factors, guides cellular responses and interactions in COVID-19 (Figure 3 ).
Figure 3.
Soluble mediators during the disease course of COVID-19
Summarizing the principles established by many observational studies addressing the role of soluble mediators of the innate immune system. In red color severe (critical and fatal) disease courses, in yellow mild to moderate disease courses. (1) The term “days from symptom onset” is increasingly used to determine the kinetics of the disease. For this term, it needs to be considered that there exists variance in interpretation as well as in the time from infection to first symptoms, which is illustrated by the fading colors in the graph. (2 and 3) Temporal phases of COVID-19 have not been defined equally in all studies. However, in the majority of studies, “early” (2) describes the period up to 10 days after onset of symptoms, and “late” (3) the period from approximately days 11–25 after onset of symptoms. (4) Admission to hospital had been used initially, because it is a concrete data point. However, the time from viral infection to hospital admission can vary significantly, which complicates comparability of longitudinal studies using admission to hospital as the starting point. (5) Viral detection levels over time summarized following findings reported in Lucas et al. (2020). (6) The early local innate immune responses are very difficult to capture in a clinical setting since the time of viral infection is not known for most patients. Evidence from model systems indicate that type I and III IFN responses are most important; however, the magnitude and exact kinetic of this response in severe versus mild COVID-19 is still under debate. (7) Local innate immune responses (mainly in upper and lower respiratory tract). In addition to chemokines, some reports also mention proinflammatory mediators. Data for later time points are still rare. (8–10) Most often mentioned soluble markers in plasma or serum of patients with mild (8), moderate to severe (9) disease, or critically ill COVID-19 patients (including fatal outcomes) (10), mainly at the end of the first week and the beginning of the second week, at the end of the first week and the beginning of the second week, and at later time points, respectively. Asymptomatic SARS-CoV-2 infections have been shown to exhibit lower levels of cytokines (Long et al., 2020). Depending on study design and methodology, not all markers have been measured in all studies and at all time points.
Early on, it was suggested that COVID-19, particularly severe disease courses, might be associated with a maladapted induction of an immune response to the infection leading to what has been previously termed a “cytokine storm” or cytokine release syndrome (CRS) (Mulchandani et al., 2021; de la Rica et al., 2020). Summarizing many observational studies, a current model suggests that SARS-CoV-2 enters type II pneumocytes via ACE2 in the respiratory system leading to rapid virus replication as well as concomitantly to a proinflammatory state with elevated levels of cytokines such as IL-1, IL-6, CXCL8, and TNF (Cao, 2020; Lucas et al., 2020; Wang et al., 2020a). This finding was recapitulated in vitro in monocyte-derived macrophages, but not DCs, suggesting that these cells might be a major source of the proinflammatory cytokines (Yang et al., 2020a). Proinflammatory genes upregulated in innate immune cells in severe or critical patients mainly belonged to the NF-κB pathway (Hadjadj et al., 2020). Accumulation of NF-κB-dependent proinflammatory mediators can provoke an accumulation of pathogenic inflammatory neutrophils and macrophages in the lung, further perpetuating higher proinflammatory cytokines and chemokines in BALF (CCL2, CCL3, CCL4, and CXCL10) (Xiong et al., 2020) but also in the circulation (IL-1, IFNγ, IL-17, TNF, IP-10, MCP-1, G-CSF, GM-CSF, IL-1RA, CCL2, CCL3, CCL5, CCL8, CXCL2, CXCL8, CXCL9, and CXCL16) (Blanco-Melo et al., 2020; Hadjadj et al., 2020; Huang et al., 2020; Wang et al., 2020a; Wu and Yang, 2020; Yang et al., 2020b). This was particularly prominent during the second week after disease onset and more pronounced in severe patients (Lucas et al., 2020), resulting in an excessive inflammatory and immune response, especially in the lungs, leading to acute respiratory distress syndrome (ARDS), pulmonary edema, apoptosis of epithelial cells, and finally vascular damage and multi-organ failure (Berlin et al., 2020; Gandhi et al., 2020). The elevation of cytokines in severe disease is accompanied by elevated clinical laboratory parameters including alanine aminotransferase, lactate dehydrogenase, C-reactive protein (CRP), ferritin, and D-dimers. Bioactivity of some of these mediators of inflammation, particularly IL-6 and TNF, has been validated by transcriptomics in blood-derived cells, demonstrating corresponding elevated IL-6 and TNF response signatures in circulating immune cells (Hadjadj et al., 2020). Inflammatory cell death in this hyperinflammatory state has been attributed to the concomitant elevation of TNF and IFNγ as being sufficient to trigger pyroptosis, apoptosis, and necrosis (PANoptosis) driving tissue damage and mortality in severe COVID-19 (Karki et al., 2021). Simultaneous upregulation of IFNγ and IL-10 with higher IFNγ levels, particularly in mild patients, is another hallmark of COVID-19 (Hadjadj et al., 2020).
The term cytokine storm has been quickly used to describe the immunopathology in severe COVID-19. Critical voices have since raised concerns whether cytokine storm or CRS reflects the immune response in COVID-19 patients correctly (Sinha et al., 2020), noting that the observed elevated IL-6 levels are considerably lower than e.g., in septic shock or CRS due to other causes (Kox et al., 2020; Monneret et al., 2021). Yet, longitudinal profiling also revealed dynamic changes in concentration of many cytokines including IL-6 (Lucas et al., 2020), highlighting the importance of temporal examination of the pathological features of this new disease. On the other hand, the definition of cytokine storm has expanded to conditions beyond sepsis and, irrespective of absolute cytokine concentrations, to include all inflammatory conditions with elevated circulating cytokines that result in systemic inflammation as well as secondary organ dysfunction (Fajgenbaum and June, 2020; Mangalmurti and Hunter, 2020), including COVID-19. One further hallmark of immune hyperactivation in cytokine storm is the failure of resolution of the inflammation (Fajgenbaum and June, 2020). Considering the coexistence of CRS in severe disease with immunosuppressive innate immune cells (Remy et al., 2020; Schulte-Schrepping et al., 2020), the exact terminology of immunopathology in severe COVID-19 still warrants further investigation. Complex viral sepsis with immune deviation has been proposed as a current alternative description (Riva et al., 2020).
Children often experience no or only mild symptoms on SARS-CoV-2 infection, but can, in rare cases, present with multisystem inflammatory syndrome in children (MIS-C) 1–2 months after infection. Symptoms such as elevated inflammatory markers, fever, and multi-organ dysfunction bear overlap to acute, severe COVID-19 in adults or also Kawasaki disease, a pediatric vasculitis, however, MIS-C has distinct immunological features different from the two with respect to circulating cytokine profiles and composition of the T cell compartment (Carter et al., 2020; Consiglio et al., 2020; Gruber et al., 2020). Although the pathogenesis is still unknown, the appearance of autoantibodies has been reported (Consiglio et al., 2020; Gruber et al., 2020).
Apart from the classical chemokines and cytokines, soluble mediators present in patients’ circulation have been investigated in omics approaches to identify systemic effectors indicating novel aspects of severe COVID-19 pathology and to find novel biomarkers and/or therapeutically targetable factors. Proteomic and metabolomic studies in plasma also revealed molecules previously reported to be linked to immunosuppression, such as kynurenines, are elevated in severe disease whereas numerous lipids were reduced (Shen et al., 2020; Su et al., 2020). Whether kynurenines and other potentially immunosuppressive molecules indeed exert such function in COVID-19 requires further functional testing. The profound loss of lipids, especially sphingolipids, glycerophospholipids, cholines, and derivatives, might contribute to loss of immune function of innate immune cells, particularly in monocyte-derived cells, such as signal transduction, growth regulation, cytokine secretion, cell migration, adhesion, apoptosis, senescence, and inflammatory responses. Proteins involved in fatty acid metabolism such as FETUB and CETP, known to suppress inflammation, were reduced in serum, as was PI6, a suppressor of chemotaxis (Shu et al., 2020). Another important class of mediators is the acute phase proteins (APPs), which are involved in early states of immune responses to viral infections. In addition to CRP and the alarmins S100A8/A9 (Schulte-Schrepping et al., 2020; Silvin et al., 2020), the serum amyloids A-1 (SAA1), A-2 (SAA2), and A-4 (SAA4), the serum amyloid P-component (SAP/APCS), and alpha-1-antichymotrypsin (SERPINA3) were found to be elevated in severe COVID-19 (Shen et al., 2020; Shu et al., 2020). Similarly, the elevation of components and regulators of the complement system (complement 6, complement factor B, properdin, and carboxypeptidase N catalytic chain) point toward complement activation as an early innate immune activation mechanism (Shen et al., 2020). In addition to thrombocytopenia, the platelet-derived chemokines proplatelet basic protein (PPBP; also called macrophage-derived growth factor) and platelet factor 4 (PF4) showed reduced levels in COVID-19 (Shen et al., 2020). Another interplay between metabolite regulation and immune mediators in COVID-19 might occur due to interferon-mediated reduction of amino acid metabolism (Shen et al., 2020).
Collectively, all these findings support a much more complex host-pathogen interaction, particularly in severe COVID-19, which is better summarized as viral sepsis, with the upregulation of proinflammatory mediators being only one part of a derailed innate immune response.
Changes in the innate immune cell compartment
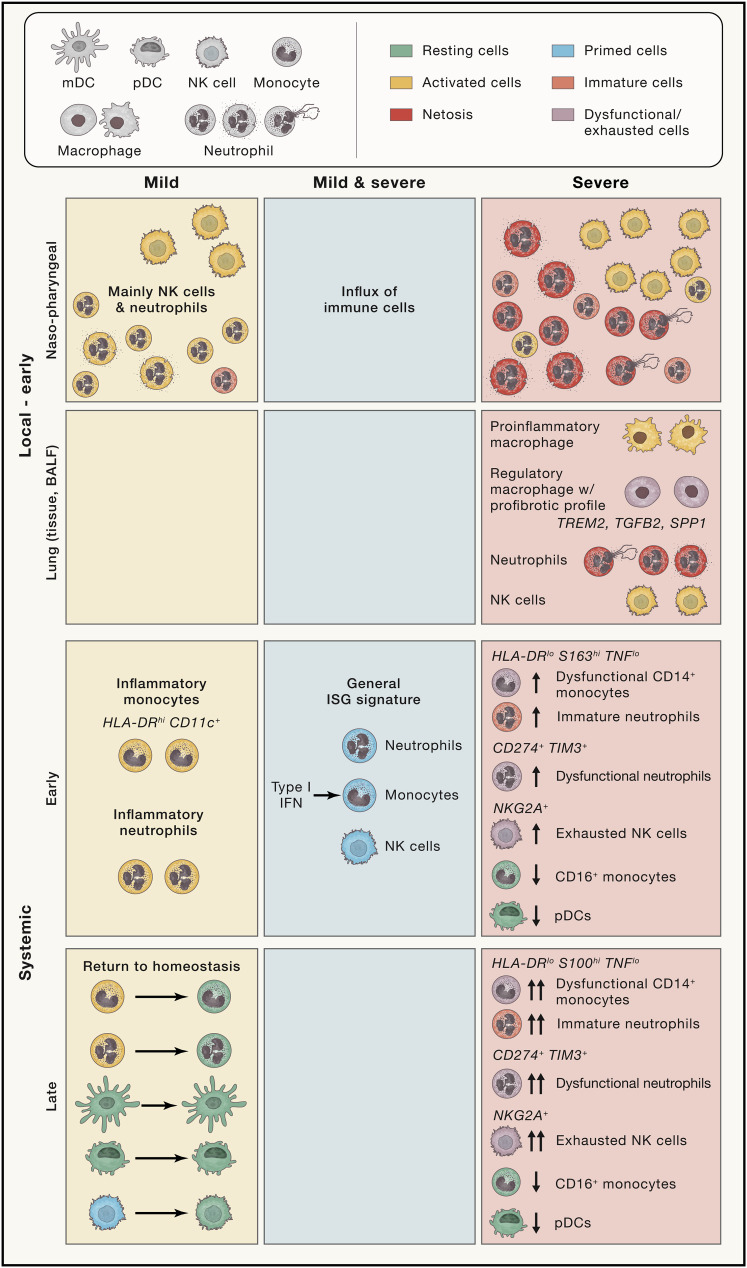
Recognition of the viral infection by tissue-resident immune cells results in a local innate immune response leading to the recruitment of further innate immune cells with the intent to initiate viral clearance (Figure 4 ). Handling of the infection in the upper respiratory tract (the major entry point of SARS-CoV-2), managing viral clearance, and consequent resolution of the active immune response, is a key aspect for prevention of viral dissemination into the lung and pulmonary injury (Newton et al., 2016).
Figure 4.
Cellular changes during COVID-19
Depicted are the COVID-19-associated changes for the innate immune cells stratified by common disease characteristics (middle column) or such associated with either mild or severe disease courses, as well as stratified by location (local versus systemic) and time. ISG, interferon response gene; Mac, macrophage; mDC, myeloid dendritic cell; NKG2A, NK cell receptor (CD159a) encoded by killer cell lectin lectin-like receptor C1, KLRC1; pDC, plasmacytoid dendritic cell; SPP1, secreted phosphoprotein 1 (coding for osteopontin); TGF-B2, transforming growth factor beta 2; TIM3, T cell immunoglobulin and mucin domain-containing 3 (also known as hepatitis A virus cellular receptor 2, HAVCR2); TREM2, triggering receptor expressed on myeloid cells 2.
The local mucosal immune response to SARS-CoV-2 infection has been characterized by cellular and transcriptional changes in the upper respiratory tract (Chua et al., 2020) as well as in BALF assessing the lower, more distal parts of the lung (Liao et al., 2020). COVID-19 patients, in general, showed the expected influx of innate immune cells, particularly neutrophils and monocytes into the nasopharyngeal mucosa on chemokine secretion of the infected epithelial cells (e.g., CXCL1, CXCL3, CXCL6, CXCL15, CXCL16, and CXCL17). Severe patients expressed elevated levels of chemokines and chemokine receptors and exhibited a marked increase in neutrophils in the tissue (Chua et al., 2020). Further, proinflammatory macrophages were identified in the lungs of critical patients to possibly contribute to excessive inflammation by promoting further granulocyte and monocyte recruitment and differentiation (Chua et al., 2020; Liao et al., 2020). Because no further functional analysis was performed, it remains to be determined whether these activated infiltrating monocyte-derived macrophages in addition to recruited neutrophils in critically ill patients are capable of exerting typical functions of phagocytic cells at the viral entry sites and/or to what extent they may contribute to tissue damage. Furthermore, because immune cells in circulation were not studied from the same patients, it also remains to be seen whether changes observed in the circulation are reflective of patient-individual changes in organs with known high viral load and infection such as the lung.
Elevated neutrophil and natural killer (NK) cell numbers, as well as lower proportions of dendritic cells and T cells, were a feature of BALF from severe patients, whereas clonally expanded CD8+ T cells were found in such samples from moderate patients (Liao et al., 2020). Further dissection of the increased monocyte/macrophage compartment in severe patients revealed macrophage subpopulations showing immunoregulatory features indicative of resolution, yet also showing expression of the profibrotic genes TREM2, TGFB2, and SPP1 (Liao et al., 2020).
The initial local respiratory SARS-CoV-2 infection entails alterations in the circulating cells in the blood with particular changes in the innate compartment. An increase in circulating neutrophils versus a decrease in lymphocytes has been appreciated as a hallmark of severe COVID-19 (Hadjadj et al., 2020; Mathew et al., 2020; Qin et al., 2020; Schulte-Schrepping et al., 2020).
Mild COVID-19 patients were marked by circulating HLA-DR hi CD11c hi inflammatory monocytes with an interferon-stimulated gene signature, while noticeable amounts of circulating neutrophil precursors appeared in severe cases, indicative of emergency myelopoiesis, as well as the presence of dysfunctional, CD274 (PD-L1)-expressing mature neutrophils (Schulte-Schrepping et al., 2020). Further, loss of non-classical monocytes as well as the appearance of dysfunctional S100hi classical monocytes with downregulated MHC class II expression were associated with severe COVID-19, reminiscent of immunoparalysis seen in sepsis (Arunachalam et al., 2020; Giamarellos-Bourboulis et al., 2020a; Kuri-Cervantes et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020; Su et al., 2020; Wilk et al., 2020). Further, elevated levels of megakaryocytes and erythroid cells were identified in severe COVID-19, and the associated expression modules correlated with mortality (Bernardes et al., 2020). Neutrophils in COVID-19 patients showed expression profiles associated with activation and neutrophil extracellular trap (NET) formation (Aschenbrenner et al., 2021; Schulte-Schrepping et al., 2020) as well as increased NET formation ex vivo (Middleton et al., 2020). These findings go hand in hand with the thrombotic complications and coagulopathy frequently found in severe COVID-19 (Klok et al., 2020), because NET formation can seed immunothrombosis (Veras et al., 2020), and platelets and neutrophils can interact to promote thrombus formation (Pircher et al., 2019). Interestingly, plasmacytoid DC (pDCs), the main source of IFNα, were not only reduced in abundance in the circulation (Kuri-Cervantes et al., 2020) but also impaired in function in COVID-19 patients suggesting the lung as a source for the elevated plasma levels of type I IFNs early on infection (Arunachalam et al., 2020; Lucas et al., 2020). Single-cell transcriptomics of circulating innate immune cells strongly support a general IFN response in all COVID-19 patients with a strong temporal component (Lee et al., 2020; Lucas et al., 2020; Schulte-Schrepping et al., 2020; Wilk et al., 2020).
In addition to an increase in circulating neutrophils, lymphopenia has been identified as a hallmark of severe COVID-19 (Guan et al., 2020; Huang et al., 2020), and an elevated neutrophil/lymphocyte ratio has been introduced as a biomarker of disease severity (Kuri-Cervantes et al., 2020; Lagunas-Rangel, 2020; Liu et al., 2020; Qin et al., 2020). Although abundance and functional changes in the B and T cell compartment are not part of this review and are discussed in greater detail elsewhere (Sette and Crotty, 2021), NK cells are affected as well (Giamarellos-Bourboulis et al., 2020a; Hadjadj et al., 2020; Kuri-Cervantes et al., 2020; Zheng et al., 2020a). Reduced abundance of circulating NK cells was noticed in severe COVID-19 cases as well as an increased surface expression of NKG2A associated with a functionally exhausted phenotype, producing lower levels of cytokines on stimulation usually necessary to fight virus-infected cells (Zheng et al., 2020a). Increased levels of cytotoxicity (PRF1), DNA replication (DDIT4), and decreased inhibition of NF-κB signaling (COMMD6) in NK cells was associated with recovery from COVID-19 (Su et al., 2020). In early studies on patients during convalescence, an increased frequency of NK cells was reported (Ni et al., 2020; Rodriguez et al., 2020).
Concerning the less frequent granulocyte subsets in circulation, elevated levels of eosinophils were found in severe COVID-19 (Lucas et al., 2020) as well as an early transient increase in a CD62L+ eosinophil subpopulation, reminiscent of lung eosinophils, albeit the potential role of these proinflammatory cells in the development of ARDS, and sudden clinical deterioration in severe patients still warrants further assessment (Rodriguez et al., 2020). Basophils were found to be reduced in COVID-19 (Laing et al., 2020), which has been noted to correlate with the humoral response (Rodriguez et al., 2020).
Immune deviations in the adaptive immune system observed in severe COVID-19 might be directly linked to the immunosuppressive phenotypes observed in innate immune cells, such as HLA-DRlo monocytes impeding proper antigen presentation, or the expression of PD-L1 on suppressive neutrophil subpopulations possibly supporting lymphopenia by inducing apoptosis of lymphocytes (Schulte-Schrepping et al., 2020). Further, the lack of costimulatory molecules on conventional DCs may contribute to functional impairment of T cell activation leading to delayed receptor binding domain- and nucleocapsid protein-specific T cell responses (Zhou et al., 2020b). Interestingly, cellular immune dysregulation of innate immune cells, particularly monocytes and DCs, seems to be sustained during convalescence irrespective of disease severity (Files et al., 2021; Zhou et al., 2020b). Further, because basophils have been implicated in potentiating the humoral immune response (Denzel et al., 2008), this might constitute another avenue of innate-adaptive crosstalk in COVID-19.
All in all, a scenario emerges with rather differently programmed innate immune cells in mild versus severe disease courses (Figure 4), which is also accompanied with similar differences in the adaptive immune response to SARS-CoV-2 (Sette and Crotty, 2021). Following the initial local innate immune response, the occurrence of a systemic immune response component seems to be independent of disease severity, albeit cellular changes differ dramatically between mild and severe cases. Further, the decision, which kind of innate immune response to this virus will occur, seems to be made very early after infection. As a consequence, follow-up studies in larger cohorts determining molecular mechanisms explaining the different responses and clinical outcomes are urgently needed, because these will likely uncover novel druggable targets to prevent severe, critical, and fatal disease courses. However, we postulate that different molecular mechanisms might all converge toward a detrimental clinical path.
Factors that influence disease severity
Genetic factors impacting the immune response to SARS-CoV-2
The clinical observation that otherwise healthy young individuals are among fatal cases suggested that host genetics might contribute to disease susceptibility and severity (Ovsyannikova et al., 2020). Several international consortia (COVID-19 host genetics initiative [COVID-19 Host Genetics Initiative, 2020] or the COVID Human Genetic Effort [Casanova et al., 2020]) address genetic susceptibility either by genome-wide association studies (GWASs) or whole genome sequencing (WGS). It was postulated that signaling pathways identified to infer susceptibility for other viral infections might also be of importance for COVID-19 (Ovsyannikova et al., 2020). An early GWAS conducted in Spain and Italy identified a gene cluster on chromosome 3 as a genetic susceptibility locus in patients with COVID-19 with respiratory failure (Severe Covid-19 GWAS Group et al., 2020). These findings were confirmed by the COVID-19 Host Genetics Initiative (COVID-19 Host Genetics Initiative, 2020) and the Genetics of Mortality in Critical Care (GenOMICC) GWAS (Pairo-Castineira et al., 2020). The COVID-19 risk locus on chromosome 3 harbors immune genes including CCR9, CXCR6, XCR1, CCR1, and CCR2. Comparative genomics strongly suggested that this COVID-19 risk locus is inherited from Neanderthals and is carried by around 50% of people in South Asia and around 16% of people in Europe (Zeberg and Pääbo, 2020). CCR1 is a receptor for several chemokines (CCL3, CCL5, CCL7, and CCL23), some of which show altered expression with COVID-19 severity (Chua et al., 2020). Furthermore, knockout of CCR1 in mice supports that this receptor protects from excessive inflammatory response (Gerard et al., 1997) and decreases susceptibility to viruses and fungi (Blease et al., 2000). From the GenOMICC study, it was proposed that critical illness in COVID-19 is associated with genetic variance in IFNAR2 and OAS2, two genes involved in antiviral defense mechanisms, as well as DPP9, TYK2, and CCR2, three genes previously associated with host-driven inflammatory lung injury, of which IFNAR2, DPP9, and CCR2 were also revealed by a transcriptome-wide association study (Pairo-Castineira et al., 2020). It needs to be mentioned that these GWASs have not yet revealed any causative relationships, and the odds ratios were rather low, indicating that genetics might only play a minor influence at these gene loci.
The COVID Human Genetic Effort consortium sequenced genomes from patients with severe COVID-19 and compared them with mild patients. Emphasizing on genes within pathways that were associated with genetic susceptibility for other viral infections, particularly influenza, they identified 13, mainly loss-of-function mutations, associated with severe COVID-19 disease course in the TLR3, IRF7, and IRF9 pathways (Zhang et al., 2020a) (Figure 5 ). pDCs from IRF7-deficient patients produced no type I IFN when infected with SARS-CoV-2, and fibroblasts from patients with deficiencies in TLR3, IRF7, and IFNAR1 were susceptible to SARS-CoV-2 infection suggesting that cell-autonomous innate immune mechanisms were not operational. The overall estimate for such deficiencies among patients with severe disease was 3.5%. These findings strongly point toward a causal relationship between genotype and phenotype and support an important role for type I IFNs in defense against SARS-CoV-2. The role of the type I IFN system was further underlined by the identification of the presence of a B cell autoimmune phenocopy of inborn type I IFN immunodeficiency in more than 10% of patients with severe COVID-19 (Bastard et al., 2020). Autoantibodies against type I IFN were more prevalent in men, which might contribute to the excess of men among patients with severe COVID-19. The understanding of such immune-phenomena will allow better patient stratification in a precision medicine approach to tailor therapeutic options such as plasmapheresis, plasmablast depletion, or early treatment with recombinant type I IFNs for these patients.
Figure 5.
Genetic factors for COVID-19 susceptibility support the central role of the innate immune system
The studies highlighted have already identified several loss-of-function mutations in molecules involved in the recognition of SARS-CoV-2 as well as important downstream signaling molecules important for a well-orchestrated and robust type I IFN response. Genes, for which loss-of-function mutations were identified, are depicted in bold pink with a pink circle. The autoimmune phenocopy of inborn type I IFN immunodeficiency is also depicted. For abbreviations, see Figure 2 legend.
Another approach identified variants of the X-chromosomal TLR7 as genetic susceptibility factors by performing genetic variance segregation analysis in two families with young family members (<35 years) admitted to the intensive care unit (ICU) due to severe COVID-19 (van der Made et al., 2020) (Figure 5). In both families, the TLR7 mutations resulted in downregulation of downstream genes including IRF7, IFNB1, and ISG15 on stimulation of PBMCs with the TLR7 ligand imiquimod. As a consequence, production of IFNγ was also decreased.
Collectively, these findings establish TLR3, TLR7, type I, and type II IFN responses as important immune mechanisms controlling infection with SARS-CoV-2. Ongoing studies are now addressing whether the assessment of these variants can be integrated in future risk scores used to identify individuals at elevated risk of developing severe COVID-19 or utilized in clinical tests to stratify patients for preventive and novel mechanism-based therapeutic measures.
Age, inflammaging, and COVID-19
Age is the major clinical risk factor for severe, critical, and fatal COVID-19 (Richardson et al., 2020; Williamson et al., 2020). Although expression of SARS-CoV-2 entry factors, such as ACE2 and TMPRSS2, in the airway have been found to increase with age (Muus et al., 2021), there is no evidence so far that SARS-CoV-2 RNA levels differ between young, adult, and aged COVID-19 patients (Jacot et al., 2020), indicating that it is the host immune response facilitating severe disease courses in the elderly. Changes observed within the innate immune system during aging might be related to an unfavorable outcome of COVID-19. In elderly, the number of dendritic cells (including pDCs) and homeostatic alveolar macrophages are reduced and so are the levels of pattern recognition receptors (Shaw et al., 2013). This is accompanied with reduced type I IFN production combined with perturbed inflammatory cytokine responses to viral infections (Canaday et al., 2010; Molony et al., 2017). Because autoantibodies to type I IFN are found preferentially in the elderly population, this provides a causal explanation for age as the major clinical risk factor in ∼10% of cases of critical COVID-19 pneumonia (Bastard et al., 2020).
The effect of the age-associated changes seen in both the innate and the adaptive arm of the immune system have been investigated in several animal models in the context of coronavirus infection (Channappanavar and Perlman, 2020; Pence, 2020). In aged macaques (Yu et al., 2020) and hamsters (Imai et al., 2020), SARS-CoV-2 infection is associated with more severe respiratory illness and fatal pneumonia compared to young animals. From experiments with SARS-CoV, it has been postulated that particularly altered innate immune responses in aged animals contribute to severe disease courses (Smits et al., 2010; Zhao et al., 2011).
Systemically, the aged innate immune system shares another hallmark of severe COVID-19, namely increased myelopoiesis with elevated numbers of myeloid cells including neutrophils and monocytes in circulation and inflammatory macrophages in the tissues (Ho et al., 2019). For all major myeloid cells, a decline or deviation of cellular functions including migration to infectious tissues, phagocytosis, production of prostaglandins, or nitric and superoxide is well established with age (Shaw et al., 2013). For a particularly strong manifestation of all these changes, the term inflammaging was coined (Boren and Gershwin, 2004; Franceschi et al., 2017). Inflammaging is considered a chronic, sterile inflammation characterized by elevated levels of inflammatory mediators in the circulation including IL-6, TNF, CXCL8, CCL2, CRP, and PLA2GD (group IID secretory phospholipase A2), which in turn have been directly linked to reduced numbers and function of myeloid DCs and pDCs (Frasca and Blomberg, 2016). Numerous factors inducing this inflammatory state have been suggested including DNA damage, increased epigenetic variations, release of mitochondrial DNA, age-related leakage of intestinal microbiota, persistent viral infections (e.g., with cytomegalovirus), or cellular immunosenescence (Cheung et al., 2018; Cunha et al., 2020; Franceschi et al., 2017).
Using inflammaging-derived signatures, it was shown that transcriptional changes observed in immune cells from COVID-19 patients are reminiscent of the aging phenotype (Schulte-Schrepping et al., 2020; Zheng et al., 2020b). Because inflammaging is a significant risk factor for age-related inflammatory conditions including cardiovascular disease, diabetes, cancer, autoimmune conditions, or Alzheimer’s disease (Boren and Gershwin, 2004; Frasca et al., 2017), it is likely to be one of the most important molecular risk factors for COVID-19. In this setting, with defective functionality in the myeloid compartment along an increase in proinflammatory monocytes and higher baseline levels of many proinflammatory mediators such as IL-6 (Shaw et al., 2013), it can be hypothesized that the initiated innate immune response during COVID-19 exacerbates the already existing disbalance of the immune system even further toward a proinflammatory and dysfunctional state. A recent study illustrated that age has an impact on the inflammatory mediators present during COVID-19 (Angioni et al., 2020). Here, CXCL8, IL-10, IL-15, IL-27, and TNF positively correlated with age, longer hospitalization, and more severe disease courses. Based on the identification of large numbers of dysfunctional immune cells, we postulate that the higher severity in aged individuals is caused by dysregulated, rather than impaired, host innate immunity.
Adaptation of innate immune cells: A role for trained immunity?
A specialized form of innate immune adaptation is the induction of immune memory (also termed trained immunity) (Netea et al., 2020a), best characterized for BCG vaccination (Giamarellos-Bourboulis et al., 2020b; Moorlag et al., 2020) and β-glucan stimulation of monocytes (Dos Santos et al., 2019; Kleinnijenhuis et al., 2012). In principle, exposure of innate immune cells and their precursors to either BCG or β-glucan leads to their reprogramming, inducing heterologous protection against other pathogens including viruses (Arts et al., 2018).
In context of COVID-19, it has been postulated that trained immunity might be a means to adapt the homeostatic state of the innate immune system toward a more favorable outcome in case of SARS-CoV-2 infection, particularly in those groups at higher risk for severe disease courses (Netea et al., 2020b; O’Neill and Netea, 2020). Early retrospective studies on the impact of BCG vaccination in childhood resulted in contradictory results concerning a beneficial role for fighting SARS-CoV-2 infection (de Chaisemartin and de Chaisemartin, 2020; Escobar et al., 2020; Hamiel et al., 2020; Kinoshita and Tanaka, 2020; Lindestam Arlehamn et al., 2020). However, more recent epidemiological observations from the first wave of the pandemic indicate that the prevalence of individuals with positive tuberculin sensitivity tests—either due to exposure to Mycobacterium spp. or BCG vaccination—inversely correlates with the incidence of COVID-19 (Singh et al., 2020). Although not providing any causal relationship for trained immunity and protection from COVID-19, such correlations triggered randomized clinical trials addressing whether trained immunity adapts the innate immune system toward a more favorable response to SARS-CoV-2 infections (Netea et al., 2020b; O’Neill and Netea, 2020). In late 2020, 21 newly initiated trials assessing protective effects of BCG vaccination against COVID-19 and 3 trials assessing VPM1002, a further development of the old BCG vaccine, were registered at https://ClinicalTrials.gov (accessed 2021/01/19) (Nowill and de Campos-Lima, 2020). First results are expected in the middle of 2021 (M. Netea, personal communication). In this context, it will also be important to see whether boosting the potential of proinflammatory cytokine production (IL-1, IL-6, and TNF) by the induction of trained immunity (Kleinnijenhuis et al., 2012) will exaggerate the observed hyperinflammatory cytokine responses in severe COVID-19, or whether trained immunity would reprogram innate immune cells, so that these cells can react more robustly during the early hours after infection thereby disrupting the path toward the vicious cycle of viral sepsis (Netea et al., 2020b; O’Neill and Netea, 2020). Further studies identifying the molecular mechanisms that could prevent reprogramming of innate immune cells toward a suppressive phenotype in severe COVID-19 are urgently needed.
Immune parameters as biomarkers
High-throughput, high-resolution, and high-content technologies assessing cellular or soluble mediators of the immune response are ideal to discover new potential biomarkers addressing important clinical questions concerning diagnostics, monitoring, outcome prediction, or therapeutic options for COVID-19. From a clinical perspective, actionable measurements of soluble factors are focused on technologies assessing serum or plasma samples, yet different combinations of soluble factors were assessed in different studies in context of COVID-19 with only a few choosing comprehensive approaches (Messner et al., 2020; Su et al., 2020).
Multi-omics analysis of plasma proteins including cytokines and chemokines distinguished mild and moderate COVID-19 (Su et al., 2020) suggesting that immune parameters might be used for disease classification or outcome prediction. In a study by the Mount Sinai Health System in New York, four cytokines (IL-6, CXCL8, TNF, and IL-1β) were assessed in serum from 1,484 COVID-19 patients (Del Valle et al., 2020) of which TNF, IL-1β, and IL-6 were elevated in COVID-19 patients, and TNF and IL-6 turned out to be independent predictors of disease severity and fatal outcome. In another study, a machine learning-based classifier of eleven plasma proteins, including orosomucoid-1/alpha-1-acid glycoprotein-1 (ORM1), orosomucoid-1/alpha-1-acid glycoprotein-2 (ORM2), fetuin-B (FETUB), and cholesteryl ester transfer protein (CETP), was generated to determine and predict disease outcome (Shu et al., 2020). Several other classifiers, including immune mediators, (Lucas et al., 2020) were developed for predicting fatality or convalescence, further indicating that immune parameters from the circulation could be utilized in the clinics. The currently ongoing and future studies need to address which of the many suggested biomarkers are best suited for diagnosis, disease outcome prediction, or therapy response. This is critical when considering the development of stratified therapies (Buszko et al., 2020).
Therapeutic options targeting immune deviation
Three major concepts emerge concerning therapeutic and preventive options in COVID-19: (1) targeting the virus and its cellular life cycle by antiviral drugs, (2) developing vaccines that can either prevent or mitigate disease symptoms after infection, and (3) targeting the deviation of the immune response to avoid or mitigate severe and fatal disease outcome (Riva et al., 2020; Vabret et al., 2020). Targeting the innate immune system might play a role in all three areas.
The antivirals are divided in direct and indirect acting antivirals, targeting molecules and mechanisms of the virus itself or host cell proteins, respectively. Because SARS-CoV-2 antagonizes the type I IFN system, drugs strengthening this cellular defense system might improve early innate immune responses. Type I IFN therapy in patients with genetic deficits in the IFN system (Zhang et al., 2020a), but not in those with autoimmune phenocopies of these deficits (Bastard et al., 2020), might be beneficial if provided sufficiently early (Hadjadj et al., 2020; Wang et al., 2020b). More than 20 clinical trials are currently evaluating the efficacy of type I IFN treatment (https://www.clinicaltrials.gov, accessed 2021/01/19) (Wang et al., 2020b; Zhou et al., 2020a), the best time window, and benefits versus risks of IFN therapy. Alternatives such as IFNλ (type III IFN) only targeting receptors on epithelial cells without the broader effects of type I IFNs (Prokunina-Olsson et al., 2020) are also under clinical evaluation. Other indirect antivirals blocking viral cell entry (e.g., by targeting proteases such as TMPRSS2) have been suggested as potential therapies (Kaur et al., 2021), yet definitive proof for clinical efficacy is still lacking. Antiviral protein interaction mapping revealed further promising sets of antivirals: those that affect translation and those that modulate the sigma-1 and sigma-2 receptors, the cellular interaction partners of SARS-CoV-2 NSP6 and ORF9c (Gordon et al., 2020).
Based on results from phase-III vaccine trials utilizing mRNA vaccines (Anderson et al., 2020; Polack et al., 2020), two vaccines have been approved in the United States, United Kingdom, Israel, and the European Union, and several million individuals have been vaccinated since late 2020. A most surprising finding in light of age-related changes in the adaptive and the innate immune system is the similarly high efficiency of these vaccines in the elderly population. This unexpected success requires further mechanistic and molecular evaluations about the elicited immune response because this might give insights how age-related alterations of the immune system are overcome by this type of vaccination.
The third strategy is targeting the deviation of the immune response to avoid or mitigate severe and fatal disease outcomes. A starting point for many therapeutic strategies has been the hyperinflammatory state in severe disease (Tang et al., 2020; Wang et al., 2020a). Not surprisingly, trials targeting cytokines such as IL-6, IL-1, IFNγ, IL-1R, TNF, CXCL8, GM-CSF, GM-CSF receptor, or IL-37 have been reported, as well as strategies attempting to achieve disruption of chemokine signaling (e.g., via CCR1, CCR2, and CCR5) to prevent overt innate immune cell recruitment into the lung (Wang et al., 2020a). Because COVID-19 presents with such heterogeneity, a certain drug might be beneficial in one setting, while having no effect in another. For example, inconsistent results have been reported from large clinical trials when trying to inhibit IL-6 associated with hyperinflammation (Huang and Jordan, 2020). A randomized clinical trial assessing tocilizumab, an anti-IL-6 antibody (Stone et al., 2020), showed no benefit for moderately ill patients concerning effectiveness of preventing intubation or death. In contrast, among critically ill patients, the risk of in-hospital mortality was lower in patients treated with tocilizumab in the first 2 days of ICU admission (Gupta et al., 2021), which has also been reported in earlier trials (Huang and Jordan, 2020). It cannot yet be ruled out that targeting IL-6 might be beneficial for a group of patients in specific clinical settings. This might be similarly true for targeting IL-1 with anakinra for which several smaller studies were reporting beneficial effects (Iglesias-Julián et al., 2020; Kooistra et al., 2020), yet results from larger randomized trials are still missing.
Whether targeting single effector molecules will be efficient in disrupting the COVID-19-associated immune deviation awaits the report of ongoing clinical trials. Targeting several of these pathways simultaneously (e.g., by inhibiting Janus kinases) might overcome such limitations (McCreary and Pogue, 2020; Wu and Yang, 2020). In early clinical trials, the use of the JAK inhibitor baricitinib showed a reduction in serum levels of IL-6, IL-1β, and TNF (Bronte et al., 2020), suppressed the production of proinflammatory cytokines in lung macrophages, and the recruitment of neutrophils to the lung (Hoang et al., 2021). Randomized clinical trials have to be initiated to validate these promising results. Along these lines, therapies targeting the kallikrein-kinin system with icatibant (van de Veerdonk et al., 2020), or the coagulation system with antibodies against C5a and C3a (Mastellos et al., 2020), have been introduced as additional options to ameliorate clinical symptoms and reduce mortality rates. Further, these therapeutic strategies might be considered in combination with immunotherapies such as anti-IL-6 or anti-IL-1 in severe COVID-19.
Reverse transcriptomics of blood immune cells derived from COVID-19 patients predicted the broadly anti-inflammatory drug dexamethasone and other corticosteroids as potentially beneficial for a subgroup of severely ill COVID-19 patients (Aschenbrenner et al., 2021). Indeed, dexamethasone was shown to be beneficial, particularly in patients with severe disease courses where it reduced 28-day mortality in randomized clinical trials (RECOVERY Collaborative Group et al., 2021; Tomazini et al., 2020; WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group et al., 2020). Further, molecular prediction of drug responses may guide the way for precision medicine approaches following patient stratification.
Summary and outlook
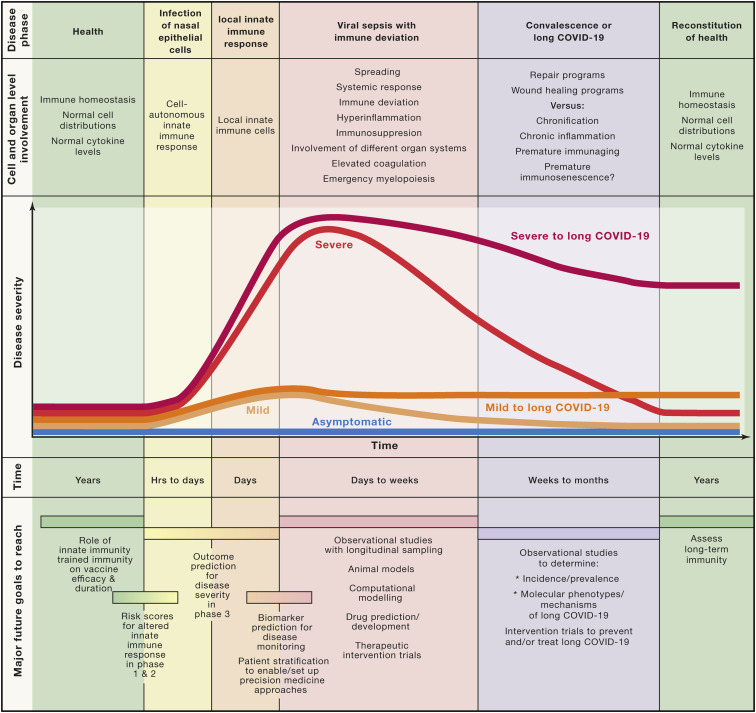
Valuable descriptive and correlative information about COVID-19 has been gathered during the first months of the pandemic (Figure 6 ). COVID-19 can be divided in at least five phases (Figure 1), starting with the infection of ACE2+ epithelial cells of the respiratory tract leading to cell-autonomous defense mechanisms of the infected cells. The second, local immune response phase is characterized by an atypical and heterogeneous type I IFN response due to SARS-CoV-2 attacking the IFN system. Further mechanistic insights into the regulation of the heterogeneity of the type I IFN response combined with the elevated expression of NF-κB-associated inflammatory genes mainly in severe COVID-19 are urgently needed. Organ involvement, magnitude of clinical symptoms, and length of this phase is highly variable, and whether the heterogeneity of innate immunity is causally related to the clinical heterogeneity also requires further work. Here, sex differences concerning innate immunity in COVID-19 need to be considered (Takahashi et al., 2020) and molecularly characterized. This is similarly true for a potential role of innate immune cells such as NK cells, innate lymphoid cells, mast cells, basophils, or eosinophils, which have not yet been studied in sufficient detail in COVID-19.
Figure 6.
Summary of current knowledge, outlook, and open questions
Established disease courses describing cell and organ involvement during different phases of COVID-19 including the possibility of not only returning to homeostasis (convalescence) but also the potential chronification of the disease (“Long COVID-19”). Possible COVID-19 disease courses are depicted as differently colored curves of disease severity over time. Major open questions and future tasks concerning the innate immune response to SARS-CoV-2 are outlined along the five phases at the bottom of the figure.
To further improve our knowledge on COVID-19 and, in particular, on innate immunity against SARS-CoV-2 infection, future studies need to be tailored to address important questions for each disease phase. To better determine individuals at risk for an incomplete innate immune response during infection and local immunity, risk scores based on genetic findings (Zhang et al., 2020a) but also integrating environmental conditions (e.g., microbiome, mucosal infections, inflammaging, immunosenescence, and comorbidities) need to be developed that would predict insufficient first line defense against SARS-CoV-2. Parameters measurable at earlier phases of the disease would be useful to predict the outcome of viral sepsis. Recent attempts have explored this in smaller studies (Bernardes et al., 2020; Del Valle et al., 2020; Shu et al., 2020; Silvin et al., 2020), however, joining forces to address some of the suggested early predictors and validate them in larger multicenter trials (Arunachalam et al., 2020; Su et al., 2020) utilizing high-content, high-throughput, high-resolution technologies is the next step to study COVID-19 systematically. A better understanding of the molecular immune mechanisms, particularly at the transition from a local to a systemic response phase and during viral sepsis, is a prerequisite to introduce novel therapies. For example, the vicious cycle of viral sepsis developing in an affected individual already suffering from chronic obstructive pulmonary disease (COPD) might differ from the disease course observed in an obese person and therefore might require other therapies. Large enough observational validation studies with longitudinal sampling will ensure sufficiently covered molecular subtypes of COVID-19, which subsequently need to be supported by mechanistic studies in animal models (Mulchandani et al., 2021).
More evidence accumulates suggesting that a yet unknown fraction of patients will suffer from what is described as Long COVID-19, a chronic illness with very heterogeneous symptoms (Marshall, 2020). These include chronic fatigue syndrome (Salisbury, 2020) and a spectrum of psychiatric disorders ranging from cognitive decline, depression, to even neurodegeneration (Taquet et al., 2021). A better description of the heterogeneity of Long COVID-19 and the underlying molecular mechanisms need to be established to provide therapeutic options for these patients. It will be interesting to see how the immune deviations seen in acute COVID-19 extend or further develop in these patients. It will be important to generate large enough registries of individuals suffering from Long COVID-19 to address the mechanisms involved in this heterogeneous syndrome.
Acknowledgments
We want to apologize to all those colleagues whose excellent work we could not reference here. A.C.A. was supported by an intramural grant from the Department of Genomics & Immunoregulation at the LIMES Institute and by an ImmunoSensation2 Women in Science grant. The work of J.L.S. was supported by the German Research Foundation (DFG) (INST 37/1049-1, INST 216/981-1, INST 257/605-1, INST 269/768-1, INST 217/988-1, INST 217/577-1, SCHU 950/8-1, GRK 2168, and TP11) and under Germany’s Excellence Strategy (DFG) (EXC2151; 390873048), the EU project SYSCID (733100), and the BMBF-funded grant iTREAT (01ZX1902B).
Declaration of interests
The authors declare no competing interests.
References
- Acharya D., Liu G., Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor S., Fernández Blanco L., Baker D. Innate immunity during SARS-CoV-2: evasion strategies and activation trigger hypoxia and vascular damage. Clin. Exp. Immunol. 2020;202:193–209. doi: 10.1111/cei.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angioni R., Sánchez-Rodríguez R., Munari F., Bertoldi N., Arcidiacono D., Cavinato S., Marturano D., Zaramella A., Realdon S., Cattelan A., et al. Age-severity matched cytokine profiling reveals specific signatures in Covid-19 patients. Cell Death Dis. 2020;11:957. doi: 10.1038/s41419-020-03151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Moorlag S.J.C.F.M., Novakovic B., Li Y., Wang S.-Y., Oosting M., Kumar V., Xavier R.J., Wijmenga C., Joosten L.A.B., et al. BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe. 2018;23:89–100.e5. doi: 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner A.C., Mouktaroudi M., Krämer B., Oestreich M., Antonakos N., Nuesch-Germano M., Gkizeli K., Bonaguro L., Reusch N., Baßler K., et al. German COVID-19 Omics Initiative (DeCOI) Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021;13:7. doi: 10.1186/s13073-020-00823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashary N., Bhide A., Chakraborty P., Colaco S., Mishra A., Chhabria K., Jolly M.K., Modi D. Single-Cell RNA-seq Identifies Cell Subsets in Human Placenta That Highly Expresses Factors Driving Pathogenesis of SARS-CoV-2. Front. Cell Dev. Biol. 2020;8:783. doi: 10.3389/fcell.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. N. Engl. J. Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- Bernardes J.P., Mishra N., Tran F., Bahmer T., Best L., Blase J.I., Bordoni D., Franzenburg J., Geisen U., Josephs-Spaulding J., et al. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity. 2020;53:1296–1314.e9. doi: 10.1016/j.immuni.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blease K., Mehrad B., Standiford T.J., Lukacs N.W., Kunkel S.L., Chensue S.W., Lu B., Gerard C.J., Hogaboam C.M. Airway remodeling is absent in CCR1-/- mice during chronic fungal allergic airway disease. J. Immunol. 2000;165:1564–1572. doi: 10.4049/jimmunol.165.3.1564. [DOI] [PubMed] [Google Scholar]
- Boren E., Gershwin M.E. Inflamm-aging: autoimmunity, and the immune-risk phenotype. Autoimmun. Rev. 2004;3:401–406. doi: 10.1016/j.autrev.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., et al. Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Bronte V., Ugel S., Tinazzi E., Vella A., De Sanctis F., Canè S., Batani V., Trovato R., Fiore A., Petrova V., et al. Baricitinib restrains the immune dysregulation in patients with severe COVID-19. J. Clin. Invest. 2020;130:6409–6416. doi: 10.1172/JCI141772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszko M., Park J.-H., Verthelyi D., Sen R., Young H.A., Rosenberg A.S. The dynamic changes in cytokine responses in COVID-19: a snapshot of the current state of knowledge. Nat. Immunol. 2020;21:1146–1151. doi: 10.1038/s41590-020-0779-1. [DOI] [PubMed] [Google Scholar]
- Callaway E. The coronavirus is mutating - does it matter? Nature. 2020;585:174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- Canaday D.H., Amponsah N.A., Jones L., Tisch D.J., Hornick T.R., Ramachandra L. Influenza-induced production of interferon-alpha is defective in geriatric individuals. J. Clin. Immunol. 2010;30:373–383. doi: 10.1007/s10875-010-9374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., et al. Brazil-UK Centre for Arbovirus Discovery, Diagnosis, Genomics and Epidemiology (CADDE) Genomic Network Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.J., Fish M., Jennings A., Doores K.J., Wellman P., Seow J., Acors S., Graham C., Timms E., Kenny J., et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. 2020;26:1701–1707. doi: 10.1038/s41591-020-1054-6. [DOI] [PubMed] [Google Scholar]
- Casanova J.-L., Su H.C., COVID Human Genetic Effort A Global Effort to Define the Human Genetics of Protective Immunity to SARS-CoV-2 Infection. Cell. 2020;181:1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chaisemartin C., de Chaisemartin L. BCG vaccination in infancy does not protect against COVID-19. Evidence from a natural experiment in Sweden. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1223. Published online August 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Age-related susceptibility to coronavirus infections: role of impaired and dysregulated host immunity. J. Clin. Invest. 2020;130:6204–6213. doi: 10.1172/JCI144115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Vallania F., Warsinske H.C., Donato M., Schaffert S., Chang S.E., Dvorak M., Dekker C.L., Davis M.M., Utz P.J., et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell. 2018;173:1385–1397.e14. doi: 10.1016/j.cell.2018.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., Tan Z., Zicari S., Ruggiero A., Pascucci G.R., et al. CACTUS Study Team The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell. 2020;183:968–981.e7. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha L.L., Perazzio S.F., Azzi J., Cravedi P., Riella L.V. Remodeling of the Immune Response With Aging: Immunosenescence and Its Potential Impact on COVID-19 Immune Response. Front. Immunol. 2020;11:1748. doi: 10.3389/fimmu.2020.01748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. Profile of a killer: the complex biology powering the coronavirus pandemic. Nature. 2020;581:22–26. doi: 10.1038/d41586-020-01315-7. [DOI] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzel A., Maus U.A., Rodriguez Gomez M., Moll C., Niedermeier M., Winter C., Maus R., Hollingshead S., Briles D.E., Kunz-Schughart L.A., et al. Basophils enhance immunological memory responses. Nat. Immunol. 2008;9:733–742. doi: 10.1038/ni.1621. [DOI] [PubMed] [Google Scholar]
- Dos Santos J.C., Barroso de Figueiredo A.M., Teodoro Silva M.V., Cirovic B., de Bree L.C.J., Damen M.S.M.A., Moorlag S.J.C.F.M., Gomes R.S., Helsen M.M., Oosting M., et al. β-Glucan-Induced Trained Immunity Protects against Leishmania braziliensis Infection: a Crucial Role for IL-32. Cell Rep. 2019;28:2659–2672.e6. doi: 10.1016/j.celrep.2019.08.004. [DOI] [PubMed] [Google Scholar]
- Escobar L.E., Molina-Cruz A., Barillas-Mury C. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19) Proc. Natl. Acad. Sci. USA. 2020;117:17720–17726. doi: 10.1073/pnas.2008410117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum D.C., June C.H. Cytokine Storm. N. Engl. J. Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Figueiredo C.S., Raony Í., Giestal-de-Araujo E. SARS-CoV-2 Targeting the Retina: Host-virus Interaction and Possible Mechanisms of Viral Tropism. Ocul. Immunol. Inflamm. 2020;28:1301–1304. doi: 10.1080/09273948.2020.1799037. [DOI] [PubMed] [Google Scholar]
- Files J.K., Boppana S., Perez M.D., Sarkar S., Lowman K.E., Qin K., Sterrett S., Carlin E., Bansal A., Sabbaj S., et al. Sustained cellular immune dysregulation in individuals recovering from SARS-CoV-2 infection. J. Clin. Invest. 2021;131:140491. doi: 10.1172/JCI140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Salvioli S., Garagnani P., de Eguileor M., Monti D., Capri M. Immunobiography and the heterogeneity of immune responses in the elderly: A focus on inflammaging and trained immunity. Front. Immunol. 2017;8:982. doi: 10.3389/fimmu.2017.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca D., Blomberg B.B. Inflammaging decreases adaptive and innate immune responses in mice and humans. Biogerontology. 2016;17:7–19. doi: 10.1007/s10522-015-9578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca D., Blomberg B.B., Paganelli R. Aging, Obesity, and Inflammatory Age-Related Diseases. Front. Immunol. 2017;8:1745. doi: 10.3389/fimmu.2017.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R.T., Lynch J.B., Del Rio C. Mild or Moderate Covid-19. N. Engl. J. Med. 2020;383:1757–1766. doi: 10.1056/NEJMcp2009249. [DOI] [PubMed] [Google Scholar]
- Gerard C., Frossard J.L., Bhatia M., Saluja A., Gerard N.P., Lu B., Steer M. Targeted disruption of the beta-chemokine receptor CCR1 protects against pancreatitis-associated lung injury. J. Clin. Invest. 1997;100:2022–2027. doi: 10.1172/JCI119734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Tsilika M., Moorlag S., Antonakos N., Kotsaki A., Domínguez-Andrés J., Kyriazopoulou E., Gkavogianni T., Adami M.E., Damoraki G., et al. Activate: Randomized Clinical Trial of BCG Vaccination against Infection in the Elderly. Cell. 2020;183:315–323.e9. doi: 10.1016/j.cell.2020.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C.N., Patel R.S., Trachtman R., Lepow L., Amanat F., Krammer F., Wilson K.M., Onel K., Geanon D., Tuballes K., et al. Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C) Cell. 2020;183:982–995.e14. doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., et al. China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- Gupta S., Wang W., Hayek S.S., Chan L., Mathews K.S., Melamed M.L., Brenner S.K., Leonberg-Yoo A., Schenck E.J., Radbel J., et al. STOP-COVID Investigators Association Between Early Treatment With Tocilizumab and Mortality Among Critically Ill Patients With COVID-19. JAMA Intern. Med. 2021;181:41–51. doi: 10.1001/jamainternmed.2020.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiel U., Kozer E., Youngster I. SARS-CoV-2 Rates in BCG-Vaccinated and Unvaccinated Young Adults. JAMA. 2020;323:2340–2341. doi: 10.1001/jama.2020.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han E., Tan M.M.J., Turk E., Sridhar D., Leung G.M., Shibuya K., Asgari N., Oh J., García-Basteiro A.L., Hanefeld J., et al. Lessons learnt from easing COVID-19 restrictions: an analysis of countries and regions in Asia Pacific and Europe. Lancet. 2020;396:1525–1534. doi: 10.1016/S0140-6736(20)32007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T.N., Pino M., Boddapati A.K., Viox E.G., Starke C.E., Upadhyay A.A., Gumber S., Nekorchuk M., Busman-Sahay K., Strongin Z., et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell. 2021;184:460–475.e21. doi: 10.1016/j.cell.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y.-H., Del Toro R., Rivera-Torres J., Rak J., Korn C., García-García A., Macías D., González-Gómez C., Del Monte A., Wittner M., et al. Remodeling of Bone Marrow Hematopoietic Stem Cell Niches Promotes Myeloid Cell Expansion during Premature or Physiological Aging. Cell Stem Cell. 2019;25:407–418.e6. doi: 10.1016/j.stem.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E., Jordan S.C. Tocilizumab for Covid-19 - The Ongoing Search for Effective Therapies. N. Engl. J. Med. 2020;383:2387–2388. doi: 10.1056/NEJMe2032071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Julián E., López-Veloso M., de-la-Torre-Ferrera N., Barraza-Vengoechea J.C., Delgado-López P.D., Colazo-Burlato M., Ubeira-Iglesias M., Montero-Baladía M., Lorenzo-Martín A., Minguito-de-la-Iglesia J., et al. High dose subcutaneous Anakinra to treat acute respiratory distress syndrome secondary to cytokine storm syndrome among severely ill COVID-19 patients. J. Autoimmun. 2020;115:102537. doi: 10.1016/j.jaut.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.M., Poly T.N., Walther B.A., Yang H.C., Wang C.-W., Hsieh W.-S., Atique S., Salmani H., Alsinglawi B., Lin M.C., et al. Clinical Characteristics and Neonatal Outcomes of Pregnant Patients With COVID-19: A Systematic Review. Front. Med. (Lausanne) 2020;7:573468. doi: 10.3389/fmed.2020.573468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot D., Greub G., Jaton K., Opota O. Viral load of SARS-CoV-2 across patients and compared to other respiratory viruses. Microbes Infect. 2020;22:617–621. doi: 10.1016/j.micinf.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur U., Chakrabarti S.S., Ojha B., Pathak B.K., Singh A., Saso L., Chakrabarti S. Targeting host cell proteases to prevent SARS-CoV-2 invasion. Curr. Drug Targets. 2021;22:192–201. doi: 10.2174/1389450121666200924113243. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Tanaka M. Impact of Routine Infant BCG Vaccination on COVID-19. J. Infect. 2020;81:625–633. doi: 10.1016/j.jinf.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A.B., Ifrim D.C., Saeed S., Jacobs C., van Loenhout J., de Jong D., Stunnenberg H.G., et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok F.A., Kruip M.J.H.A., van der Meer N.J.M., Arbous M.S., Gommers D.A.M.P.J., Kant K.M., Kaptein F.H.J., van Paassen J., Stals M.A.M., Huisman M.V., Endeman H. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., Sato K., USFQ-COVID19 Consortium SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra E.J., Waalders N.J.B., Grondman I., Janssen N.A.F., de Nooijer A.H., Netea M.G., van de Veerdonk F.L., Ewalds E., van der Hoeven J.G., Kox M., Pickkers P., RCI-COVID-19 Study Group Anakinra treatment in critically ill COVID-19 patients: a prospective cohort study. Crit. Care. 2020;24:688. doi: 10.1186/s13054-020-03364-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kox M., Waalders N.J.B., Kooistra E.J., Gerretsen J., Pickkers P. Cytokine Levels in Critically Ill Patients With COVID-19 and Other Conditions. JAMA. 2020;324:1565–1567. doi: 10.1001/jama.2020.17052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunas-Rangel F.A. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. J. Med. Virol. 2020;92:1733–1734. doi: 10.1002/jmv.25819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Shin E.-C. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.-S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lindestam Arlehamn C.S., Sette A., Peters B. Lack of evidence for BCG vaccine protection from severe COVID-19. Proc. Natl. Acad. Sci. USA. 2020;117:25203–25204. doi: 10.1073/pnas.2016733117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Du X., Chen J., Jin Y., Peng L., Wang H.H.X., Luo M., Chen L., Zhao Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Loo Y.-M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M., et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalmurti N., Hunter C.A. Cytokine Storms: Understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Netea M.G. Trained Innate Immunity, Epigenetics, and Covid-19. N. Engl. J. Med. 2020;383:1078–1080. doi: 10.1056/NEJMcibr2011679. [DOI] [PubMed] [Google Scholar]
- Marshall M. The lasting misery of coronavirus long-haulers. Nature. 2020;585:339–341. doi: 10.1038/d41586-020-02598-6. [DOI] [PubMed] [Google Scholar]
- Mastellos D.C., Pires da Silva B.G.P., Fonseca B.A.L., Fonseca N.P., Auxiliadora-Martins M., Mastaglio S., Ruggeri A., Sironi M., Radermacher P., Chrysanthopoulou A., et al. Complement C3 vs C5 inhibition in severe COVID-19: Early clinical findings reveal differential biological efficacy. Clin. Immunol. 2020;220:108598. doi: 10.1016/j.clim.2020.108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaleuskaya L., Veltrop R., Ikpeze N., Martin-Garcia J., Navas-Martin S. Protective role of Toll-like Receptor 3-induced type I interferon in murine coronavirus infection of macrophages. Viruses. 2012;4:901–923. doi: 10.3390/v4050901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCreary E.K., Pogue J.M. Coronavirus disease 2019 treatment: A review of early and emerging options. Open Forum Infect. Dis. 2020;7:ofaa105. doi: 10.1093/ofid/ofaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner C.B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., Textoris-Taube K., Vernardis S.I., Egger A.-S., Kreidl M., et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020;11:11–24.e4. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E.A., He X.-Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., Mostyka M., Baxter-Stoltzfus A., Borczuk A.C., Loda M., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molony R.D., Nguyen J.T., Kong Y., Montgomery R.R., Shaw A.C., Iwasaki A. Aging impairs both primary and secondary RIG-I signaling for interferon induction in human monocytes. Sci. Signal. 2017;10:eaan2392. doi: 10.1126/scisignal.aan2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monneret G., Benlyamani I., Gossez M., Bermejo-Martin J.F., Martín-Fernandez M., Sesques P., Wallet F., Venet F. COVID-19: What type of cytokine storm are we dealing with? J. Med. Virol. 2021;93:197–198. doi: 10.1002/jmv.26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorlag S.J.C.F.M., van Deuren R.C., van Werkhoven C.H., Jaeger M., Debisarun P., Taks E., Mourits V.P., Koeken V.A.C.M., de Bree L.C.J., Ten Doesschate T., et al. Safety and COVID-19 Symptoms in Individuals Recently Vaccinated with BCG: a Retrospective Cohort Study. Cell Rep. Med. 2020;1:100073. doi: 10.1016/j.xcrm.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Fauci A.S. Emerging Pandemic Diseases: How We Got to COVID-19. Cell. 2020;182:1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Daszak P., Taubenberger J.K. Escaping Pandora’s Box - Another Novel Coronavirus. N. Engl. J. Med. 2020;382:1293–1295. doi: 10.1056/NEJMp2002106. [DOI] [PubMed] [Google Scholar]
- Mulchandani R., Lyngdoh T., Kakkar A.K. Deciphering the COVID-19 cytokine storm: Systematic review and meta-analysis. Eur. J. Clin. Invest. 2021;51:e13429. doi: 10.1111/eci.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muus C., Luecken M.D., Eraslan G., Sikkema L., Waghray A., Heimberg G., Kobayashi Y., Vaishnav E.D., Subramanian A., Smillie C., et al. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. Nat Med. 2021 doi: 10.1038/s41591-020-01227-z. [DOI] [Google Scholar]
- Netea M.G., Domínguez-Andrés J., Barreiro L.B., Chavakis T., Divangahi M., Fuchs E., Joosten L.A.B., van der Meer J.W.M., Mhlanga M.M., Mulder W.J.M., et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020;20:375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Giamarellos-Bourboulis E.J., Domínguez-Andrés J., Curtis N., van Crevel R., van de Veerdonk F.L., Bonten M. Trained Immunity: a Tool for Reducing Susceptibility to and the Severity of SARS-CoV-2 Infection. Cell. 2020;181:969–977. doi: 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A.H., Cardani A., Braciale T.J. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin. Immunopathol. 2016;38:471–482. doi: 10.1007/s00281-016-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X., et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowill A.E., de Campos-Lima P.O. Immune Response Resetting as a Novel Strategy to Overcome SARS-CoV-2-Induced Cytokine Storm. J. Immunol. 2020;205:2566–2575. doi: 10.4049/jimmunol.2000892. [DOI] [PubMed] [Google Scholar]
- O’Neill L.A.J., Netea M.G. BCG-induced trained immunity: can it offer protection against COVID-19? Nat. Rev. Immunol. 2020;20:335–337. doi: 10.1038/s41577-020-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onabajo O.O., Banday A.R., Stanifer M.L., Yan W., Obajemu A., Santer D.M., Florez-Vargas O., Piontkivska H., Vargas J.M., Ring T.J., et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020;52:1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Nieuwenhuijse D.F., Stein M., O’Toole Á., Haverkate M., Mollers M., Kamga S.K., Schapendonk C., Pronk M., Lexmond P., et al. Dutch-Covid-19 response team Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat. Med. 2020;26:1405–1410. doi: 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G., Haralambieva I.H., Crooke S.N., Poland G.A., Kennedy R.B. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol. Rev. 2020;296:205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. Published online December 11, 2020. [DOI] [PubMed] [Google Scholar]
- Patil S., Fribourg M., Ge Y., Batish M., Tyagi S., Hayot F., Sealfon S.C. Single-cell analysis shows that paracrine signaling by first responder cells shapes the interferon-β response to viral infection. Sci. Signal. 2015;8:ra16. doi: 10.1126/scisignal.2005728. [DOI] [PubMed] [Google Scholar]
- Pence B.D. Severe COVID-19 and aging: are monocytes the key? Geroscience. 2020;42:1051–1061. doi: 10.1007/s11357-020-00213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher J., Engelmann B., Massberg S., Schulz C. Platelet-Neutrophil Crosstalk in Atherothrombosis. Thromb. Haemost. 2019;119:1274–1282. doi: 10.1055/s-0039-1692983. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Alphonse N., Dickenson R.E., Durbin J.E., Glenn J.S., Hartmann R., Kotenko S.V., Lazear H.M., O’Brien T.R., Odendall C., et al. COVID-19 and emerging viral infections: The case for interferon lambda. J. Exp. Med. 2020;217:e20200653. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N., Almouzni G., Gorski S.A., Aerts S., Amit I., Bertero M.G., Bock C., Bredenoord A.L., Cavalli G., Chiocca S., et al. LifeTime Community Working Groups LifeTime and improving European healthcare through cell-based interceptive medicine. Nature. 2020;587:377–386. doi: 10.1038/s41586-020-2715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand U., Rinas M., Schwerk J., Nöhren G., Linnes M., Kröger A., Flossdorf M., Kály-Kullai K., Hauser H., Höfer T., Köster M. Multi-layered stochasticity and paracrine signal propagation shape the type-I interferon response. Mol. Syst. Biol. 2012;8:584. doi: 10.1038/msb.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group. Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., et al. Dexamethasone in Hospitalized Patients with Covid-19 - Preliminary Report. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy K.E., Mazer M., Striker D.A., Ellebedy A.H., Walton A.H., Unsinger J., Blood T.M., Mudd P.A., Yi D.J., Mannion D.A., et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5:e140329. doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rica R., Borges M., Gonzalez-Freire M. COVID-19: In the Eye of the Cytokine Storm. Front. Immunol. 2020;11:558898. doi: 10.3389/fimmu.2020.558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., et al. the Northwell COVID-19 Research Consortium Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva G., Nasillo V., Tagliafico E., Trenti T., Comoli P., Luppi M. COVID-19: more than a cytokine storm. Crit. Care. 2020;24:549. doi: 10.1186/s13054-020-03267-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L., Pekkarinen P.T., Lakshmikanth T., Tan Z., Consiglio C.R., Pou C., Chen Y., Mugabo C.H., Nguyen N.A., Nowlan K., et al. Systems-Level Immunomonitoring from Acute to Recovery Phase of Severe COVID-19. Cell Rep. Med. 2020;1:100078. doi: 10.1016/j.xcrm.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi W., Kubota N., Shimizu T., Saruta J., Fuchida S., Kawata A., Yamamoto Y., Sugimoto M., Yakeishi M., Tsukinoki K. Existence of SARS-CoV-2 Entry Molecules in the Oral Cavity. Int. J. Mol. Sci. 2020;21:6000. doi: 10.3390/ijms21176000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury H. Helen Salisbury: When will we be well again? BMJ. 2020;369:m2490. doi: 10.1136/bmj.m2490. [DOI] [PubMed] [Google Scholar]
- Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Deutsche COVID-19 OMICS Initiative (DeCOI) Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severe Covid-19 GWAS Group. Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.C., Goldstein D.R., Montgomery R.R. Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol. 2013;13:875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T., Ning W., Wu D., Xu J., Han Q., Huang M., Zou X., Yang Q., Yuan Y., Bie Y., et al. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity. 2020;53:1108–1122.e5. doi: 10.1016/j.immuni.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., et al. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Maurya R.P., Singh R.K. “Trained immunity” from Mycobacterium spp. exposure or BCG vaccination and COVID-19 outcomes. PLoS Pathog. 2020;16:e1008969. doi: 10.1371/journal.ppat.1008969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P., Matthay M.A., Calfee C.S. Is a “Cytokine Storm” Relevant to COVID-19? JAMA Intern. Med. 2020;180:1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- Smits S.L., de Lang A., van den Brand J.M.A., Leijten L.M., van IJcken W.F., Eijkemans M.J.C., van Amerongen G., Kuiken T., Andeweg A.C., Osterhaus A.D.M.E., Haagmans B.L. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6:e1000756. doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.H., Frigault M.J., Serling-Boyd N.J., Fernandes A.D., Harvey L., Foulkes A.S., Horick N.K., Healy B.C., Shah R., Bensaci A.M., et al. BACC Bay Tocilizumab Trial Investigators Efficacy of Tocilizumab in Patients Hospitalized with Covid-19. N. Engl. J. Med. 2020;383:2333–2344. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C.L., Voillet V., Duvvuri V.R., Scherler K., Troisch P., et al. ISB-Swedish COVID19 Biobanking Unit Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell. 2020;183:1479–1495.e20. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., et al. Yale IMPACT Research Team Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talemi S.R., Höfer T. Antiviral interferon response at single-cell resolution. Immunol. Rev. 2018;285:72–80. doi: 10.1111/imr.12699. [DOI] [PubMed] [Google Scholar]
- Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Luciano S., Geddes J.R., Harrison P.J. Bidirectional associations between COVID-19 and psychiatric disorder: retrospective cohort studies of 62 354 COVID-19 cases in the USA. Lancet Psychiatry. 2021;8:130–140. doi: 10.1016/S2215-0366(20)30462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazini B.M., Maia I.S., Cavalcanti A.B., Berwanger O., Rosa R.G., Veiga V.C., Avezum A., Lopes R.D., Bueno F.R., Silva M.V.A.O., et al. COALITION COVID-19 Brazil III Investigators Effect of Dexamethasone on Days Alive and Ventilator-Free in Patients With Moderate or Severe Acute Respiratory Distress Syndrome and COVID-19: The CoDEX Randomized Clinical Trial. JAMA. 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Sinai Immunology Review Project Immunology of COVID-19: Current State of the Science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk F.L., Kouijzer I.J.E., de Nooijer A.H., van der Hoeven H.G., Maas C., Netea M.G., Brüggemann R.J.M. Outcomes Associated With Use of a Kinin B2 Receptor Antagonist Among Patients With COVID-19. JAMA Netw. Open. 2020;3:e2017708. doi: 10.1001/jamanetworkopen.2020.17708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F.P., Pontelli M.C., Silva C.M., Toller-Kawahisa J.E., de Lima M., Nascimento D.C., Schneider A.H., Caetité D., Tavares L.A., Paiva I.M., et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217:e20201129. doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jiang M., Chen X., Montaner L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D., et al. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell Host Microbe. 2020;28:455–464.e2. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., Weizman O.-E., Liu F., Dai Y., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021 doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group. Sterne J.A.C., Murthy S., Diaz J.V., Slutsky A.S., Villar J., Angus D.C., Annane D., Azevedo L.C.P., Berwanger O., et al. Association Between Administration of Systemic Corticosteroids and Mortality Among Critically Ill Patients With COVID-19: A meta-analysis. JAMA. 2020;324:1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers F., Subedi N., van Buuringen N., Heister D., Vivié J., Beeren-Reinieren I., Woestenenk R., Dolstra H., Piruska A., Jacobs J.F.M., et al. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat. Commun. 2018;9:3317. doi: 10.1038/s41467-018-05784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Ekiert D.C., Krause J.C., Hai R., Crowe J.E., Jr., Wilson I.A. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Chu H., Hou Y., Chai Y., Shuai H., Lee A.C.-Y., Zhang X., Wang Y., Hu B., Huang X., et al. Attenuated Interferon and Proinflammatory Response in SARS-CoV-2-Infected Human Dendritic Cells Is Associated With Viral Antagonism of STAT1 Phosphorylation. J. Infect. Dis. 2020;222:734–745. doi: 10.1093/infdis/jiaa356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127.e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., Bao L., Deng W., Gao H., Xiang Z., et al. Age-related rhesus macaque models of COVID-19. Animal Model. Exp. Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeberg H., Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. COVID-STORM Clinicians. COVID Clinicians. Imagine COVID Group. French COVID Cohort Study Group. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort. NIAID-USUHS/TAGC COVID Immunity Group Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu Y., Wang X., Yang L., Li H., Wang Y., Liu M., Zhao X., Xie Y., Yang Y., et al. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J. Hematol. Oncol. 2020;13:120. doi: 10.1186/s13045-020-00954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Legge K., Perlman S. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Invest. 2011;121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Liu X., Le W., Xie L., Li H., Wen W., Wang S., Ma S., Huang Z., Ye J., et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell. 2020;11:740–770. doi: 10.1007/s13238-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Chen V., Shannon C.P., Wei X.-S., Xiang X., Wang X., Wang Z.-H., Tebbutt S.J., Kollmann T.R., Fish E.N. Interferon-α2b Treatment for COVID-19. Front. Immunol. 2020;11:1061. doi: 10.3389/fimmu.2020.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K.-W., Wong Y.-C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.-Y., Lau T.T.-K., et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]