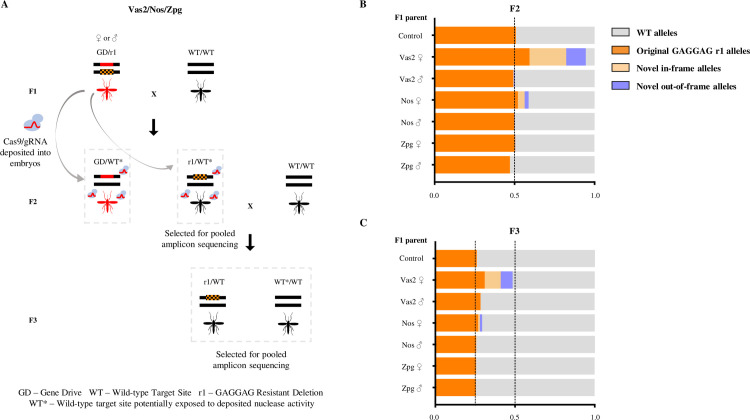
Fig 3. Heritability and rates of formation of mutations caused by parentally deposited nuclease from different gene drive constructs.
(A) zpg-CRISPRh, nos-CRISPRh or vas2-CRISPRh were crossed to a strain homozygous for a defined r1 resistance allele (203-GAGGAG) to generate F1 heterozygotes of genotype GD/r1, containing both the gene drive allele (red bar) and the resistant allele (orange hatched bar). F1 heterozygotes and r1/r1 homozygotes (control) were crossed to wild type and their non-drive F2 progeny analysed by pooled amplicon sequencing across the target site in AGAP007280. The non-drive F2 progeny (r1*/wt) were crossed to wild type, and the entire F3 progeny was analysed by amplicon sequencing to determine the heritability of mutations caused by paternally-derived nuclease. (B) Amplicon sequencing results from F2 individuals (n>128) with genotype WT/r1 (selected by absence of dsRED-linked gene drive allele), whose only source of Cas9 nuclease would be from parental deposition into the embryo. In the absence of any deposited source of nuclease only the original r1 allele and the WT allele are expected, at a ratio of 1:1. Significant deviations from this ratio, or the presence of new target site mutations, therefore indicate the mutagenic activity of deposited Cas9. (C) Amplicon sequencing of F3 progeny deriving from those F2 crossed to wild type, in order to determine the heritability of mutations formed by parentally deposited nuclease. Mendelian inheritance of mutations present in the sampled F2 individuals would be expected to lead to a 2-fold reduction in their frequency between the F2 and F3 samples.