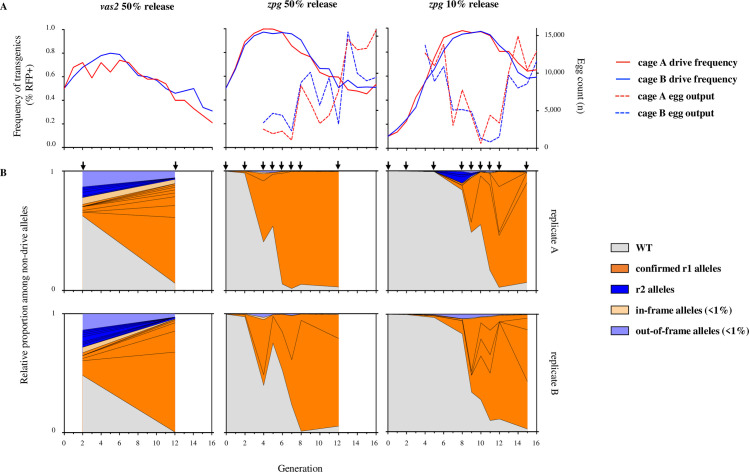
Fig 5. zpg-CRISPRh rates of spread through caged populations, reproductive load and real-time profiling of resistance.
zpg-CRISPRh/+ were released into replicate caged WT populations at 10% (right) or 50% (middle) CRISPRh/+ initial frequency. (A) The frequency of drive-modified mosquitoes (solid line) was recorded each generation by screening larval progeny for presence of DsRed linked to the CRISPRh allele. Entire egg output per generation was also recorded (dashed line). zpg-CRISPRh drive spread was compared to previous results for vas2-CRISPRh (left panel) from Hammond et al. (2017). (B) The nature and frequency of non-drive alleles was determined for several generations (arrows) of the caged population experiments by amplicon sequencing across the target site at AGAP007280 in pooled samples. Vertically aligned panels represent the two replicates from each treatment. The frequency of all WT alleles is shown in grey. Mutant alleles individually exceeding 1% frequency in any generation were labelled as ‘confirmed r1 alleles’ (orange) or ‘r2 alleles’ (blue), all had been previously confirmed as r1 or r2. Mutations that were individually present at less than 1% frequency were grouped together as in-frame (light orange) or out-of-frame (light blue).