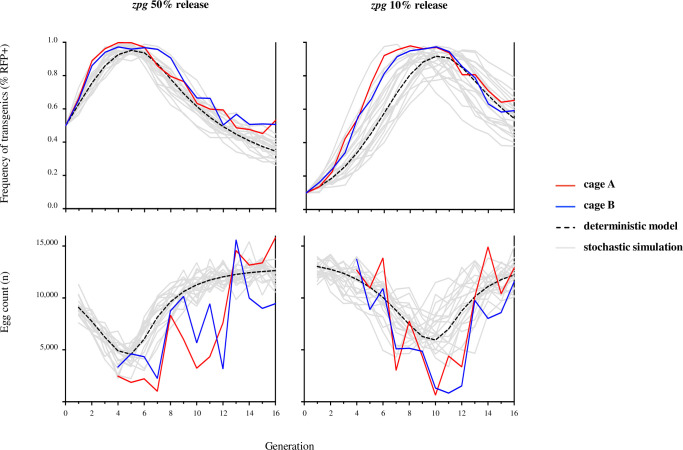
Fig 6. Comparison of observed zpg-CRISPRh drive frequency and egg output data to deterministic and stochastic model predictions.
zpg-CRISPRh/+ individuals were released into replicate caged WT populations at 50% (left) or 10% (right) initial frequency of CRISPRh/+. The frequency of drive-modified mosquitoes (top panels) and the entire egg output per generation (bottom panels) were recorded. Observed data were compared to a deterministic, discrete-generation model (dashed line) based upon observed rates of fertility, homing, and resistance generated by end-joining in the germline and embryo (S1 Table). Grey lines show 20 stochastic simulations assuming that females may fail to mate, or mate once with a male of a given genotype according to its frequency in the male population; mated females produce eggs in amounts determined by sampling with replacement from experimental values that subsequently hatch according to the genotype of the mother; of the 600 larvae chosen randomly to populate next generation, some fail to survive to adulthood. Probabilities of mating, egg production, hatching and survival from larval stage to adult are estimated from experiment (S2 Table) and random numbers for these events drawn from the appropriate multinomial distributions. Model egg counts for each release are normalised such that the deterministic value corresponds to the average of the two replicate cage experiments at generation sixteen.