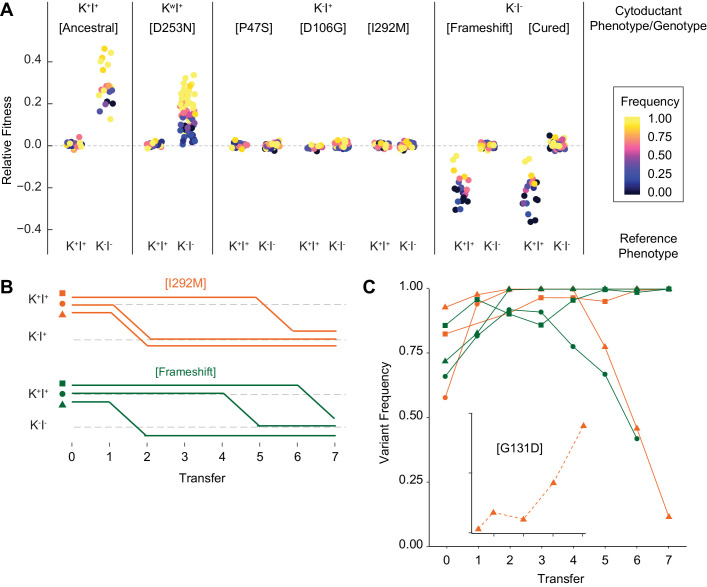
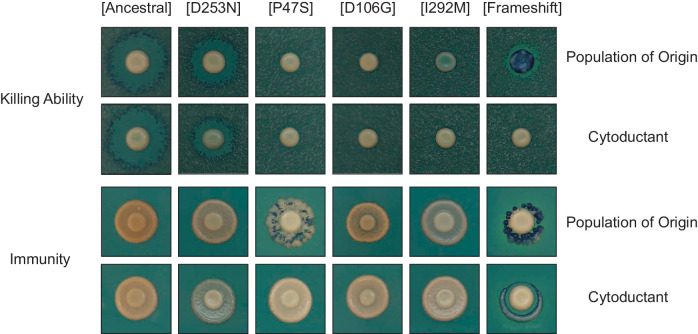
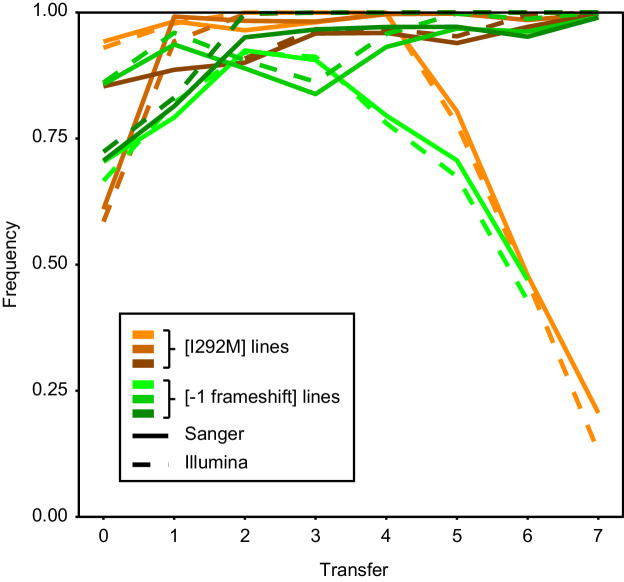
Figure 5. Viral evolution is driven by selection for an intracellular competitive advantage.
(A) Relative fitness of viral variants in pairwise competition with the ancestor (K+I+) and virus-cured ancestor (K-I-). Killer phenotype and identity of viral variant labeled above (Kw indicates weak killing ability). Killer phenotype of the ancestral competitor labeled below. Starting frequency indicated by color. (B) Change to killer phenotype during intracellular competitions between viral variants (by color) and ancestral virus. Replicate lines indicated by symbol. (C) Variant frequency during intracellular competitions. Colors and symbols consistent with panel B. Inset: frequency of the de novo G131D viral variant.