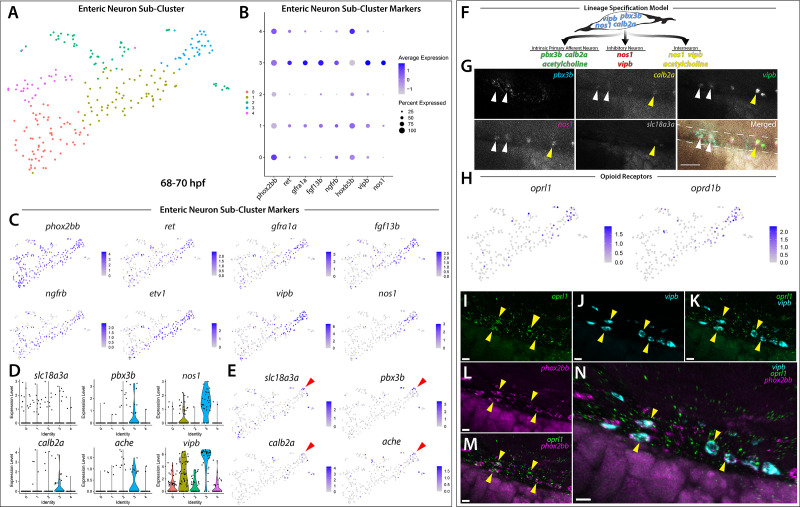
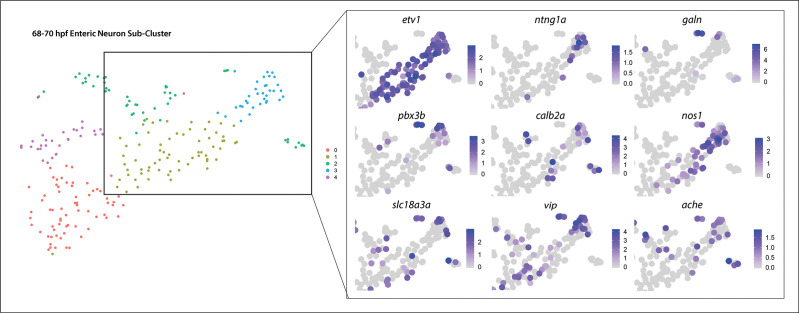
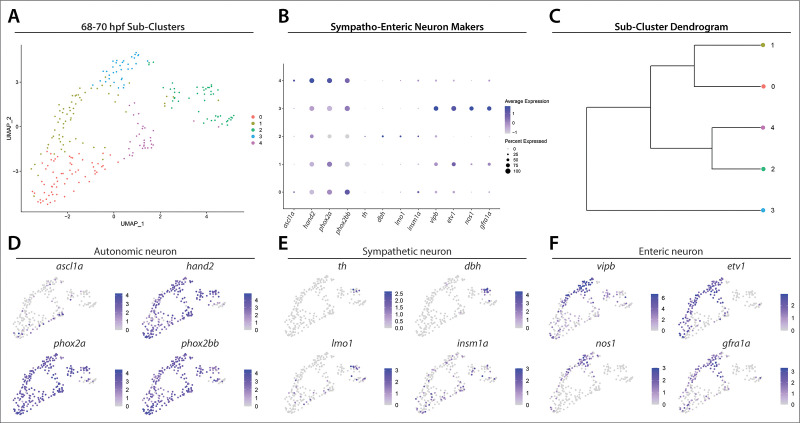
Figure 5. Differentiating enteric neurons captured during key transitional stage of subtype diversification within 68–70 hpf sox10:GFP+ larval cells.
(A) tSNE plot reveals five distinct sub-clusters following the subset analysis and re-clustering of Clusters 5 and 12 from the 68–70 hpf data set. (B) Dot plot depicts expression levels of enteric neuron markers across resulting Sub-clusters. Each marker was expressed at low levels in Sub-cluster 1 and were found to be expressed at higher levels within Sub-cluster 3. (C) tSNE feature plots further depict the expression of enteric neuron markers by illustrating the levels and localization of expression within the Sub-cluster architecture. Feature plots supplement dot plot and demonstrate the prominent expression of enteric neuron markers within Sub-cluster 3, which appears to emanate from Sub-cluster 1. (D,E) Violin and feature plots reveal expression levels of acetylcholine-associated and excitatory neuron markers reported to distinguish enteric IPANs. These markers were found in a discrete pocket of cells forming the distal-most region of Sub-cluster 3 (red arrowhead). Violin data points depicted in each Sub-cluster represent single cells expressing each gene shown. (F) Graphical model summarizes expression patterns observed in 68–70 hpf data set and HCR validation. Common enteric neuroblast capable of diverging into subsequent lineages, IPAN, inhibitory neuron, and interneuron through lineage restricted gene expression. pbx3b promotes assumption of IPAN role through loss of nos1 and vipb and begins expressing calb2a, ache, and slc18a3a. (G) Whole mount HCR analysis reveals co-expression of IPAN marker genes, pbx3b and calb2a, and inhibitory neurochemical marker genes, vipb and nos1 (white arrowheads), within the foregut (dashed white line) at 68 hpf. Vesicular acetylcholine transferase, slc18a3a, was not observed in tandem with pbx3b but was co-expressed with calb2a, vipb, and nos1 (yellow arrowheads). Scale bar: 50 μM. (H) Feature plots reveal expression of opioid receptor genes, oprl1 and oprd1b, within the differentiated enteric neuron Sub-cluster 3. (I–N) Whole mount HCR analysis validates expression of oprl1 in combination with vipb and phox2bb (yellow arrowheads) in enteric neurons localized to the foregut region of a 68 hpf embryo. Scale bar: 10 μM.