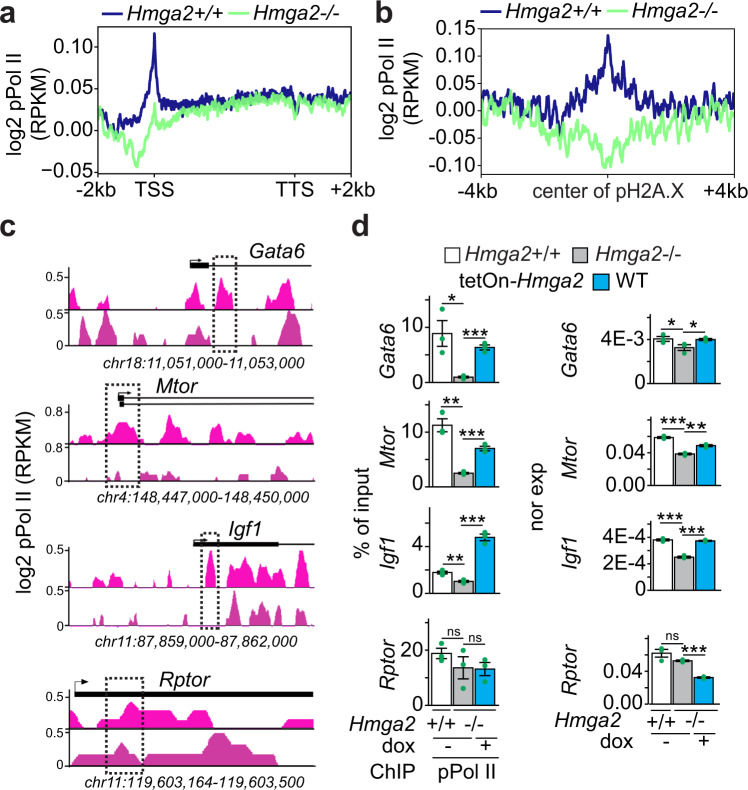
Fig. 2. Position of transcription initiating S5 phosphorylated RNA polymerase II at the TSS is Hmga2-dependent.
a, b Aggregate plots for phosphorylated serine 5 RNA polymerase II (pPol II) enrichment within the gene body ±2 kb of UCSC Known Genes (a) and in a ± 4 kb region respective to pH2A.X peaks (b) in Hmga2+/+ and Hmga2−/− MEF. ChIP-seq reads were normalized using reads per kilobase per million (RPKM) measure and are represented as log2 enrichment over their corresponding inputs. TSS, transcription start site; TTS, transcription termination site. c Visualization of selected HMGA2 target genes using UCSC Genome Browser showing pPol II enrichment in Hmga2 + /+ and −/− MEF. ChIP-seq reads were normalized using RPKM measure and are represented as log2 enrichment over their corresponding inputs. Images represent the indicated gene loci with their genomic coordinates. Arrows, direction of the genes; black boxes, exons; dotted squares, regions selected for single gene analysis. d Analysis of selected HMGA2 target genes. Left, ChIP of Gata6, Mtor, Igf1 and Rptor after pPol II immunoprecipitation in Hmga2 + /+ and Hmga2−/− MEF. Right, qRT-PCR-based, Tuba1a-normalized expression analysis under the same conditions. Bar plots presenting data as means; error bars, s.e.m (n = 3 biologically independent experiments); asterisks, P-values after two-tailed t-test, ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05. See also Supplementary Figs. 1–2 . Source data are provided as a Source Data files 01 and 04.