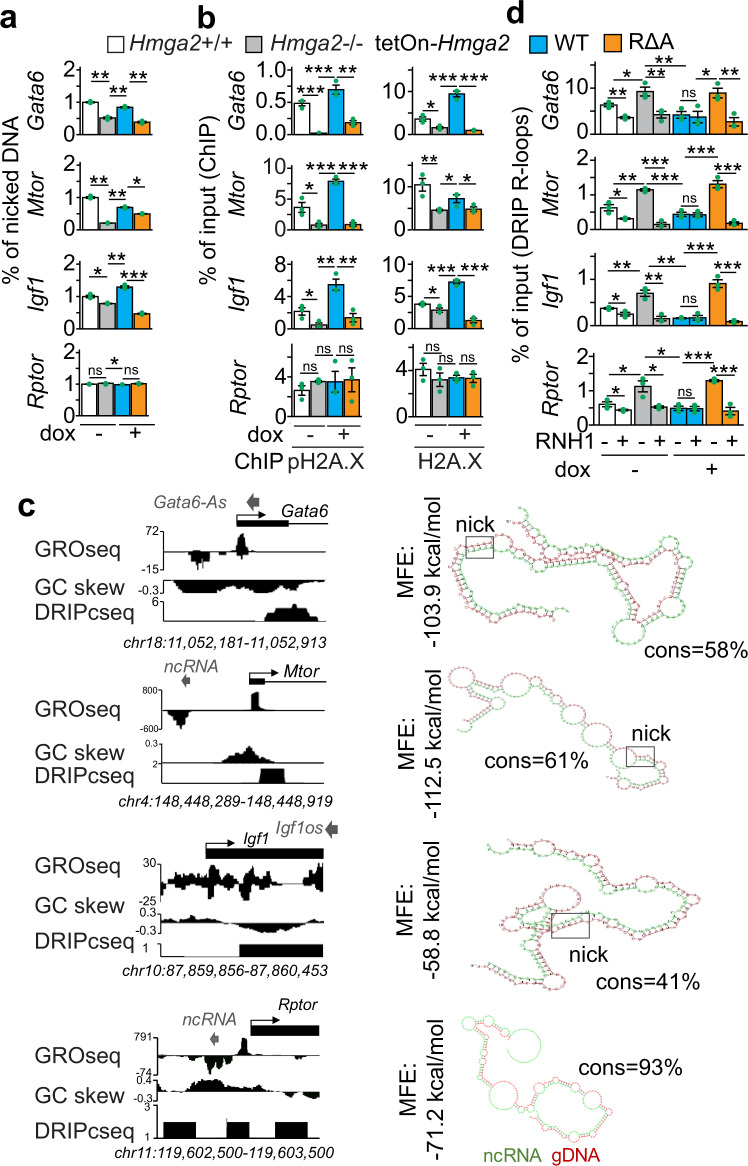
Fig. 6. HMGA2-lyase activity is required for pH2A.X deposition and solving of R-loops.
a Analysis of single-strand DNA breaks at promoters of selected HMGA2 target genes using genomic DNA from Hmga2 + /+, Hmga2−/− MEF, as well as Hmga2−/− MEF that were stably transfected with a tetracycline-inducible expression construct (tetOn) either for WT Hmga2-myc-his or the lyase-deficient mutant RΔA Hmga2-myc-his. MEF were treated with doxycycline as indicated. b ChIP-based promoter analysis of selected HMGA2 target genes using the indicated antibodies and chromatin from MEF as in a. c Left, Genome-browser visualization of selected HMGA2 target genes showed nascent RNA (GRO-seq) in WT MEF68, GC-skew and RNA-seq on plus-strand after DNA-RNA hybrid immunoprecipitation (DRIPc-seq) in NIH/3T3 mouse fibroblasts40. Images represent mapped sequence tag densities relative to the indicated loci. Genomic coordinates are shown at the bottom. Arrow heads, non-coding RNAs in antisense orientation; Arrows, direction of the genes; black boxes, exons. Right, in silico analysis revealed complementary sequences between the identified antisense ncRNA (green) and genomic sequences at the TSS of the corresponding mRNAs (red) with relatively favorable minimum free energy (MFE) and high percentage of complementarity (cons), supporting the formation of DNA-RNA hybrids containing a nucleotide sequence that favors DNA nicks (squares). d Analysis of selected HMGA2 target genes by DNA–RNA immunoprecipitation (DRIP) using the antibody S9.641 and nucleic acids isolated from MEF treated as in a. Prior IP, nucleic acids were digested with RNase H1 (RNH1) as indicated. In all bar plots, data are shown as means ± s.e.m. (n = 3 biologically independent experiments); asterisks, P-values after two-tailed t-Test, ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; ns, non-significant. See also Supplementary Figs. 5–7. Source data are provided as a Source Data file 01.