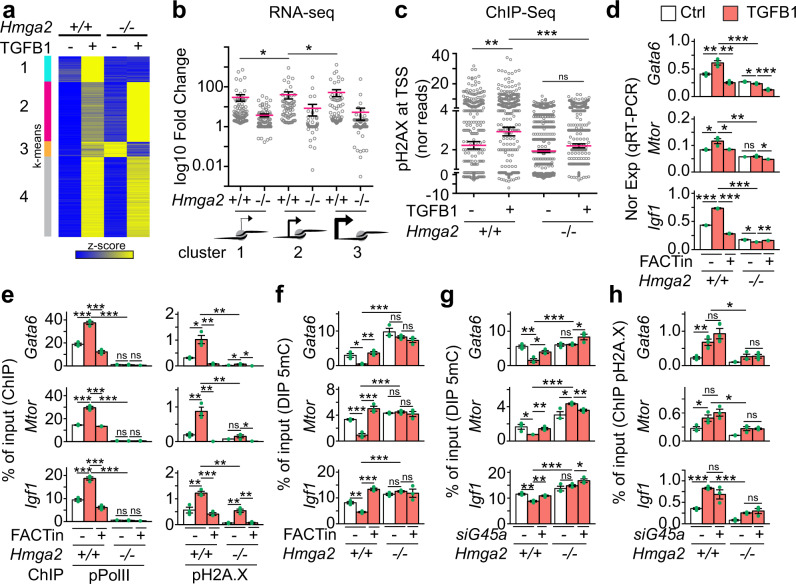
Fig. 8. HMGA2-FACT-ATM-pH2A.X axis mediates TGFB1 induced transcription activation.
a Heat map showing RNA-seq-based expression analysis of TGFB1-inducible genes in Hmga2 + /+ and Hmga2−/− MEF non-treated or treated with TGFB1. Data were normalized by Z score transformation and clustered using k-means algorithm. b Dot plot presenting the RNA-seq-based expression analysis of cluster 1 genes from A as log10 fold change between TGFB1-treated and non-treated MEF. Genes were grouped into the position clusters identified in Fig. 3a. Each dot represents the value of a single gene; n = 95 genes in position cluster 1; n = 64 genes in position cluster 2; n = 78 genes in position cluster 3; red line, average; error bars, s.e.m.; asterisks, P-values after one-tailed Mann–Whitney Test, *P ≤ 0.05. c Dot plot presenting ChIP-seq-based pH2A.X enrichment analysis in the top 15% candidates from Fig. 1c using chromatin from MEF treated as in a. For each gene, normalized reads in the TSS + 0.25 kb region were binned and the maximal value was plotted. Inputs were subtracted from the corresponding samples. Red line, average; n = 640 genes; error bars, s.e.m.; asterisks, P-values after one-tailed Mann–Whitney test, ***P ≤ 0.001; **P ≤ 0.01; ns, non-significant. d qRT-PCR-base expression analysis of HMGA2 target genes in Hmag2 + /+, Hmag2−/− MEF that were non-treated (Ctrl) or treated with TGFB1 and FACT inhibitor (FACTin; CBLC000 trifluoroacetate) as indicated. e ChIP-based promoter analysis of selected HMGA2 target genes using the indicated antibodies and chromatin from MEF treated as in d. f DIP-based promoter analysis of selected HMGA2 target genes using antibodies specific for 5-methylcytosine (5mC) and genomic DNA from MEF treated as in d. g DIP-based promoter analysis of selected HMGA2 target genes using the indicated antibodies and genomic DNA from Hmag2 + /+ or Hmag2−/− MEF that were transfected with control (−) or Gadd45a-specific small interfering RNA (siRNA) as indicated. h ChIP-based promoter analysis of selected HMGA2 target genes using pH2A.X-specific antibodies and chromatin from MEF treated as in g. In all bar plots, data are shown as means ± s.e.m. (n = 3 biologically independent experiments); asterisks, P-values after two-tailed t-test, ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; ns, non-significant. See also Supplementary Fig. 9. Source data are provided as a Source Data files 01 and 04.