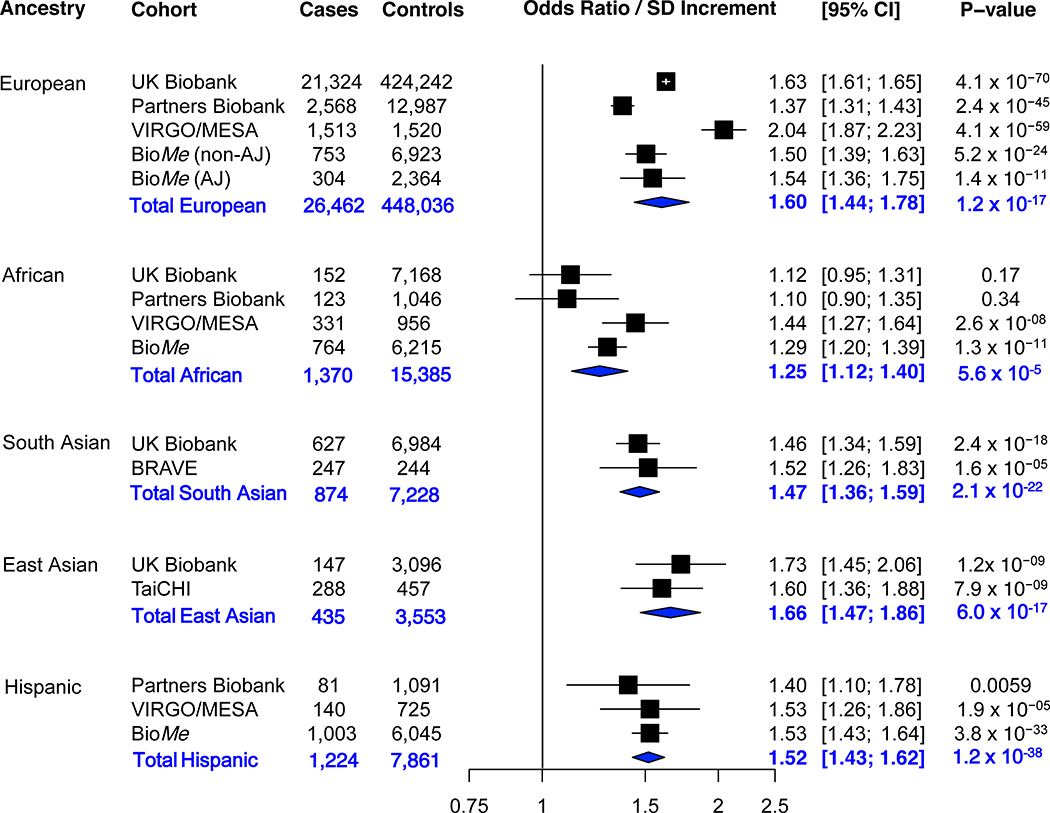
Figure.
Association of a genome-wide polygenic score with coronary artery disease in five ancestry groups. In each cohort, the polygenic score is residualized to the first four principal components of ancestry (to adjust for the effect of ancestry on the score). In each ancestry of each study, the odds ratio per standard deviation is calculated using a logistic regression model adjusted for the first four principal components of ancestry (to adjust for the effect of ancestry on disease). Sensitivity analyses adjusting for sex and the first four principal components of ancestry, or age, sex, and the first four principal components of ancestry demonstrated similar results. A random-effects meta-analysis of odds ratio per standard deviation increment was performed. The p-value for subgroup differences was 0.005 (Cochran-Mantel-Haenszel test). In pairwise ancestry group comparisons, the effect estimate in African ancestry group was significantly lower than each of the other ancestry groups, European (p=0.002), South Asian (p=0.02), East Asian (0.001), and Hispanic (0.003). None of the other ancestry group pairwise comparisons were statistically significant (p>0.05). The random effects model results for each ancestry group showed an I2 of 95.4%, 63.7%, 0%, 0%, and 0% for European, African, South Asian, East Asian, and Hispanic, respectively. Abbreviations: AJ Ashkenazi Jewish, CI confidence interval, SD standard deviation.