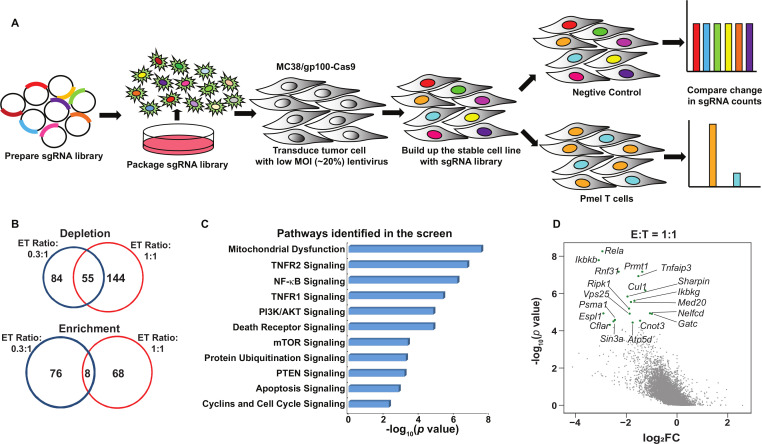
Figure 1.
Systematical discovery of tumor intrinsic factor regulating sensitivity to T cell-mediated killing. (A) A schematic diagram of the functional genome-wide CRISPR/Cas9-based immune screen strategy. A genome-wide sgRNA library (five sgRNAs per gene) was transduced into Cas9-expressing MC38/gp100 cells. Pooled MC38/gp100 cells were cocultured with tumor-reactive Pmel T cells, and followed by the next-generation sequencing of sgRNA representation. Tumors without T cell treatment were served as controls. (B) A Venn diagram illustrating the degree of overlapped candidates identified by two screens under different selection pressure. The numbers of genes whose gRNAs were significantly depleted or enriched in T cell-treated groups (|log2 (fold-change)|>1 and p<0.05) at the effector and target ratio (ET) of 0.3:1 (blue) and/or 1:1 (red) were indicated. (C) Ingenuity pathway analysis of results from the screen at the ET ratio of 1:1 revealed tumor-associated immunosuppressive pathways. (D) A scatterplot of individual gene changes in the T cell-treated groups at the ET ratio of 1:1. The changes of genes were plotted based on fold changes of their targeting sgRNAs and p values computed by MAGeCK (Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout) when compared the groups with and without T cell treatment. Top depleted genes in T cell-treated groups were highlighted with green dots. MOI, multiplicity of infection.