Abstract
The recent outbreak of coronavirus disease 2019 (COVID-19) is highly infectious, which threatens human health and has received increasing attention. So far, there is no specific drug or vaccine for COVID-19. Therefore, it is urgent to establish a rapid and sensitive early diagnosis platform, which is of great significance for physical separation of infected persons after rapid diagnosis. Here, we propose a colorimetric/SERS/fluorescence triple-mode biosensor based on AuNPs for the fast selective detection of viral RNA in 40 min. AuNPs with average size of 17 nm were synthesized, and colorimetric, surface enhanced Raman scattering (SERS), and fluorescence signals of sensors are simultaneously detected based on their basic aggregation property and affinity energy to different bio-molecules. The sensor achieves a limit detection of femtomole level in all triple modes, which is 160 fM in absorbance mode, 259 fM in fluorescence mode, and 395 fM in SERS mode. The triple-mode signals of the sensor are verified with each other to make the experimental results more accurate, and the capacity to recognize single-base mismatch in each working mode minimizes the false negative/positive reading of SARS-CoV-2. The proposed sensing platform provides a new way for the fast, sensitive, and selective detection of COVID-19 and other diseases.
Keywords: COVID-19, AuNPs, Colorimetric, SERS, Fluorescence, Triple-mode biosensor
Graphical abstract
1. Introduction
A causative coronavirus, since then named severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2), outbroke in Wuhan, Hubei Province, China, in December 2019 [1,2]. The World Health Organization (WHO) officially named 2019 novel coronavirus disease (COVID-19) [3,4], with estimated incubation period of about 2–10 days [5]. The new pneumonia caused by SARS-CoV-2 virus is highly infectious disease, and the number of infections worldwide is currently increasing. Early diagnosis of COVID-19 is very important, not only to improve the survival rate of patients, but also to hold back the infection in time. As a result, it is urgent to develop a rapid, sensitive and accurate diagnosis methods in order to effectively identify these early infects.
Current testing approaches for SARS-CoV-2 detecting can be divided into two categories, serological and nucleic acid. Serological assay is mainly to detect antibodies produced by individuals as result of exposure to the virus or antigenic proteins in infected individuals [6]. The determination of SARS-CoV-2 exposure relies largely on the detection of either IgM or IgG antibodies, for instance, using enzyme-linked immunosorbent assay (ELISA) [7] or field-effect transistors (FET) based biosensing platform [8]. However, the production of IgG and IgM antibodies in patients, termed seroconversion, usually occurs 5–10 days after the initial symptoms appear [9], therefore, nucleic acid-based testing is currently the common-used tool for early detection of SARS-CoV-2 infection. For RNA virus infections, RT-PCR [10,11] is routinely used approach for COVID-19 diagnosis, LAMP [12] and metagenomics sequencing [13,14] are also newly developed methods to diagnose COVID-19.
Recently, in order to detect biomolecules with high sensitivity, a number of novel sensing approaches have been developed by using colorimetry [15,16], fluorescence, SERS [17,18], electrochemical [19,20], and surface plasmon resonance (SPR) [21,22]. Each platform has its specific advantages, but it is still challenging to solve the problem of false negative detection, which is extremely important in epidemic prevention and control. Therefore, the method based on multiple signal output provides a powerful choice that can greatly reduce false negative signals and improve the accuracy of detection. For example, the combination of colorimetry and fluorescence have achieved highly sensitive detection of alkaline phosphatase [23], SO2 derivatives [24]; the fluorescence-SERS dual-signal platform has accomplished selective detection of microRNA [25], hydroxyl radicals [26] and K+ [27]. The colorimetrical and SERS dual-signal sensor hasve achieved precise detection of mercury ion [28] and the electrochemical-photoelectrochemical dual-mode sensing platform has accomplished the selective detection of hydrogen sulfide [29]. However, most multiple platforms are confined in dual-mode sensing, and there are still much room to fabricate the multi-mode sensing platform to meet high-demand clinical sensing.
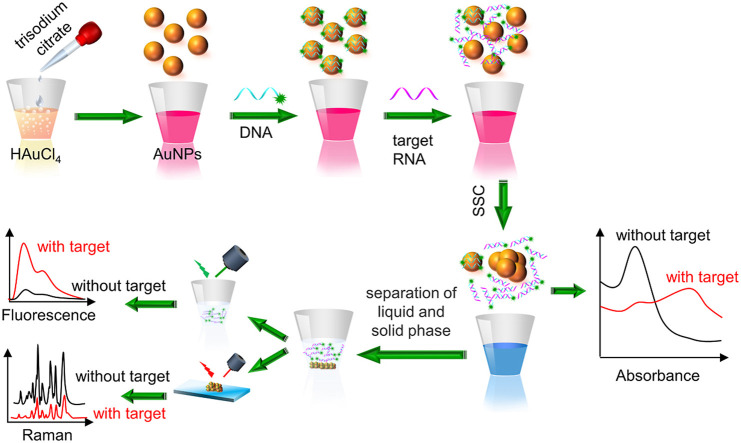
Here, we develop a triple-mode biosensor based on AuNPs to detect specific RNA in SARS-CoV-2 virus including colorimetric, SERS, and fluorescence. As shown in Fig. 1 , Citrate-stabilized AuNPs are well dispersed against aggregation due to the negative capping agent’s electrostatic repulsion. Once saline sodium citrate (SSC) is added, salt-induced self-aggregation of AuNPs results in a significant decrease in absorbance and presents visible color change. When AuNPs are mixed with appropriate concentration of DNA probes, probes get adsorbed on the surface of AuNPs and protect them against the SSC induced aggregation. In the presence of target RNAs, DNA specifically bind with target RNA forming DNA-RNA complex, which is then released from the surface of AuNPs, and the unprotected AuNPs aggregate with introduction of SSC. The target RNA concentration is transduced into the shift and intensity change of absorbance peak, the fluorescence intensity variation of the supernatant and Raman signal change of AuNPs conjugated with residue DNA probes. Each working mode in the proposed sensor is capable of recognizing single-base mismatch in target gene, which minimizes false negative/positive readings. The extraction and purification of viral RNA is not necessary for the proposed biosensor. Comparing with PCR based detection, the proposed biosensor has very simple detection process. DNA probe conjugated reaction solution can be stored in the reaction chamber and get ready for detection because of its long-term stability, the operator only needs to load detection sample and then separate the supernatant from aggregated AuNPs through centrifugation after addition of SSC buffer. The fluorescence intensity is tested by photoluminescence system and the absorption spectrum is tested by the automatic and portable microplate reader, while the Raman spectrum is recorded by a Micro Raman spectrometer. The proposed detection approach could detect 96 or 384 samples simultaneously using a 96 or 384 microplate.
Fig. 1.
Schematic of the triple-mode biosensors for COVID-19 virus RNA detection.
2. Materials and methods
2.1. Materials and apparatus
Among the SARS-CoV-2 genomic regions, it is discovered that three regions had conserved sequences: the RNA-dependent RNA polymerase gene (RdRp) in the open reading frame ORF1ab region, the envelope (E) genes and the nucleocapsid (N) protein genes [30]. Both the RdRP and E genes had high analytical sensitivity for detection [6], therefore, we selected specific fragments of ORF1ab and E genes [31] in the sequence of Wuhan-Hu-1 strain (GenBank accession number MN908947) for virus recognition based on the specific gene segment. The sequence of the DNA probe is complementary to the RNA sequence. DNA probes and RNA were ordered from Shanghai Bio-engineering Co. (Shanghai, China). The sequences of DNA and RNA were presented in Table 1 . TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and 20 × SSC (saline sodium citrate, 300 mM trisodium citrate, 3 M NaCl, pH = 7.4) were purchased from Solarbio Science and Technology Corporation, Beijing, China. Tetrachloroauric acid (HAuCl4·4H2O) and sodium citrate were ordered from Sinopharm Chemical Reagent Co.
Table 1.
Sequence information of the oligonucleotides.
| Description | Name | Sequence (5′ to 3′) |
|---|---|---|
| DNA probe | P-ORF1 | GTGAAATGGTCATGTGTGGCGGTTCACTATATGTTAAACCAGGTGGAAC-Cy3 |
| P-ORF2 | CTCATCAGGAGATGCCACAACTGCTTATGCTAATAGTGTTTTTAACATTT-Cy3 | |
| P-E1 | ACAGGTACGTTAATAGTTAATAGCGTACTTCTTTTTCTTGCTTTCGTGGTATTCTT-Cy3 | |
| P-E2 | GCTAGTTACACTAGCCATCCTTACTGCGCTTCGATTGTGTGCGTACTGCTGCAATA-Cy3 | |
| RNA | ORF1 | GUUCCACCUGGUUUAACAUAUAGUGAACCGCCACACAUGACCAUUUCAC |
| ORF2 | AAAUGUUAAAAACACUAUUAGCAUAAGCAGUUGUGGCAUCUCCUGAUGAG | |
| E1 | AAGAAUACCACGAAAGCAAGAAAAAGAAGUACGCUAUUAACUAUUAACGUACCUGU | |
| E2 | UAUUGCAGCAGUACGCACACAAUCGAAGCGCAGUAAGGAUGGCUAGUGUAACUAGC | |
| M1-1 | AUUCCACCUGGUUUAACAUAUAGUGAACCGCCACACAUGACCAUUUCAC | |
| M1-2 | GUUCCACCUGGUUUAACAUAUAGUGAACCGCCACACAUGACCAUUUCAA | |
| M1-3 | GUUCCACCUGGUUUAACAUAUAGUAAACCGCCACACAUGACCAUUUCAC | |
| M2-1 | AUUCCACCUGGUUUAACAUAUAGUGAACCGCCACACAUGACCAUUUCAA | |
| M2-2 | GUUCCACCUGGUUUAACAUAUAGUAUACCGCCACACAUGACCAUUUCAC | |
| M2-3 | GUUCCACCUGGUUUAACAUAUUGUGAACCACCACACAUGACCAUUUCAC |
Note: P-ORF1, P-ORF2, P-E1, P-E2 are probes complementary to target RNA ORF1, ORF2, E1, and E2. M1-1, M1-2, M1-3 are RNA sequences with one mismatch with probe P-ORF1, and M2-1, M2-2, M2-3 are RNA sequences with two mismatches with probe P-ORF1.
Ultraviolet visible (UV–vis) absorption spectra were recorded on a microplate reader (BioTek Instruments, SynergyTM H1) using a transparent 96-well microplate (Corning, cat. no. 3595) in the wavelength range of 400 nm–800 nm. Transmission electron microscopy (TEM) measurements were conducted on a FEI Tecnai G2 F20 electron microscope. SERS and fluorescence analysis of the Cy3 fluorophore were conducted by a Renishaw inVia Raman microscope. The Eppendorf centrifuge is used for centrifugal separation of solution.
2.2. Systhesis of AuNPs
The AuNPs were synthesized by the reduction of HAuCl4 using trisodium citrate [32]. Briefly, an aqueous solution of HAuCl4 (0.01%, 200 ml) was heated to boiling, after which 6 ml of trisodium citrate solution (1%) was added quickly into the boiling solution with vigorously stirring. The color of solution changed from yellow to wine red. The solution was heated for 20 min additionally, and then continue stirring to cool to room temperature. The prepared AuNPs solution can be stored for a relatively long time in the dispersed state. Ultrasonic dispersion for 5 min is preferred before each use.
2.3. Tri-mode detection of target RNA
The detailed procedure for RNA detection is as the following. First, DNA probes (0.05 μM, 10 μl) were added to AuNPs solutions (2.33 nM, 85 μl) and incubated for 30 min at room temperature. After incubation, the DNA probes were adsorbed onto AuNPs completely based on the nonspecific binding. Then, different concentrations of RNA (10 μl) were added to the mixed solution and incubated for 20 min at room temperature. Finally, 5 μl 0.3 × SSC was added to induce the aggregation of AuNPs. The absorbance of the solution is measured by a microplate reader. Then, the solution was centrifuged at 10,000 rpm at 4 °C for 15 min. The supernatants were transferred and the concentration of the double strands was detected by fluorescence. The solid was collected and the concentration of DNA probes was detected by a Micro Raman spectrometer. Those results corroborated each other.
3. Result and discussion
3.1. Detection principle of the triple-mode biosensor
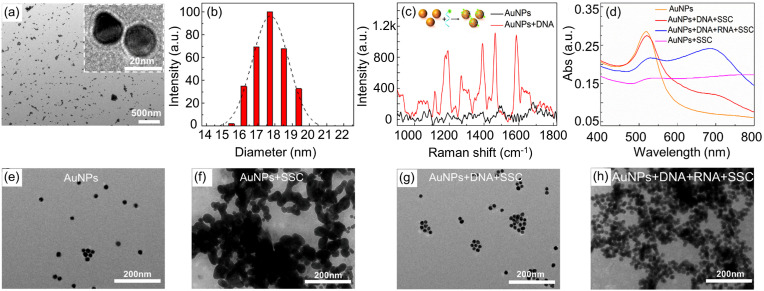
The size of AuNPs plays important role in the proposed biosensors, which determines their aggregation and the capacity to bind DNA probes [33]. AuNPs were synthesized by the reduction of HAuCl4 using trisodium citrate, and viewed under TEM, as shown in Fig. 2 a. AuNPs are spheres with similar size, and the statistical result from TEM images indicates that AuNPs have an average diameter of 17.7 nm, as in Fig. 2b. AuNPs are commonly used nanomaterials in biosensors for nucleic acid analysis, but covalent bonds [34] are usually used to immobilize DNA on the surface of AuNPs through laborious and time-consuming functionalization. In this work, single-strand DNA probes are tightly adhered to AuNPs through non-specific binding [35,36]. In order to confirm this principle, DNA probes are mixed with the prepared AuNPs and incubated for 30 min. Then, the excess DNA in supernatant is removed by centrifugation at 10,000 rpm for 15 min at 4 °C. The washing step for DNA-AuNPs conjugates is repeated two times to thoroughly remove the unadsorbed DNA, and then Raman spectrometer is used to identify the DNA probes adsorbed on AuNPs. As shown in Fig. 2c, AuNPs with DNA probes present strong representative peaks of Cy3 at 1195, 1273, 1394, 1471 and 1587 cm−1 under a laser excitation of 633 nm, indicating the immobilization of probe on AuNPs. UV–Vis spectra of AuNPs are recorded at different stages, confirming the feasibility of this method in Fig. 2d. Citrate-stabilized AuNPs have an absorption peak at ∼520 nm, once SSC is added, salt-induced self-aggregation of AuNPs results in a significant decrease in absorbance. When AuNPs are mixed with appropriate concentration of DNA, the addition of salt only slightly reduces the absorbance of AuNPs, indicating that the DNA protects AuNPs against the SSC induced aggregation. In the presence of RNA, DNA specifically binds with target RNA, and the adsorption of the DNA-RNA complex on the surface of AuNPs becomes weak, and unprotected AuNPs aggregate with introduction of SSC. As a result, the characteristic absorption peak of AuNPs at ∼520 nm decreases, and the absorption peak at 690 nm increases. At the same time, the TEM images of AuNPs clearly present their aggregation status at different stage, consistent with the UV–Vis spectra. As Fig. 2e–h shown, pure gold nanoparticles are well dispersed in deionized water, aggregate under the introduction of salt, get well dispersed again with the addition of DNA, and finally aggregate with the loading of target RNA because of the release of formed DNA-RNA complex from AuNPs. Because the RNA concentration is lower than the DNA concentration, the excess DNA still has a certain protective effect on AuNPs, so the degree of aggregation in Fig. 2h is lower than that in Fig. 2f.
Fig. 2.
(a) TEM image and (b) size distribution of the AuNPs. (c) Raman spectra of the AuNPs (black) and DNA probe (0.1 μM) conjugated AuNPs (red). (d) UV–Vis spectra of AuNPs-based biosensor. TEM images of (e) pure AuNPs (2.33 nM), (f) AuNPs aggregation induced by SCC, (g) re-dispersed AuNPs with conjugation of DNA probes (0.1 μM), and (h) re-aggregated AuNPs in the presence of target RNA (0.05 μM). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Optimization of experimental conditions
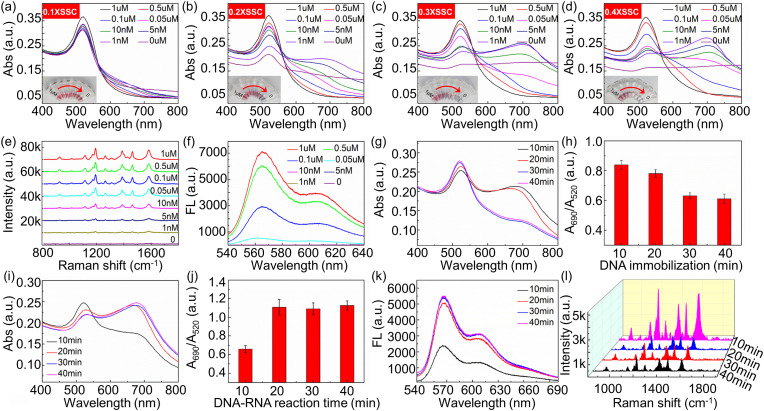
In order to achieve high-performance detection, experimental parameters are optimized including the concentration of salt, incubation time and concentration of DNA probes, and the reaction time between DNA probes and RNA. The concentration of salt has an important influence on the experiment. The stability of AuNPs is destroyed by the addition of salt due to the screening of charges on the nanoparticle surface. High concentration of salts may induce the irreversible aggregation of the nanoparticles that may sediment out of solution as a precipitate [37,38]. To investigate the effect of SSC on the aggregation behavior of AuNPs, we firstly incubate the same amount of AuNPs (85 μl) with DNA probes and then different concentration of SSC (5 μl) are added into the mixers. As shown in Fig. 3 a, when SSC with a concentration of 0.1 × is added, the concentration of DNA probes has negligible effect on the absorbance spectrum, and the color of the solution is kept in red, indicating that more SSC is needed to make AuNPs aggregate. As SSC concentration increases to 0.2 × (Fig. 3b), the absorbance of AuNPs changes significantly, but the color of the solution is mostly red when there is no DNA, indicating the concentration of SSC cannot fully aggregate AuNPs. When 0.3 × SSC is present (Fig. 3c), pure AuNPs can be aggregated completely. With the increase of DNA probes concentration, the absorbance of AuNPs changes more sensitively, and the color changes from light blue to bright red become obvious. Higher concentration of DNA is required to inhibit the aggregation of AuNPs when SSC concentration is increased to 0.4 × (Fig. 3d). Therefore, 0.3 × SSC is selected to be the optimized concentration to detect RNA in this study.
Fig. 3.
Absorption spectra of reaction system containing different concentrations of DNA probes upon the addition of 0.1 × SSC (a), 0.2 × SSC (b), 0.3 × SSC (c), 0.4 × SSC (d), respectively. inset: colorimetric changes of AuNPs/DNA/SSC. Raman spectra (e) and fluorescence spectra (f) of AuNPs containing different concentrations of DNA probes upon the addition of 0.3 × SSC. (g) Absorbance spectra and (h) A690nm/A520nm of reaction system under different incubation time between probe DNA (0.05 μM) and AuNPs. (i) Absorbance spectra, (j) A690nm/A520nm, (k) fluorescence intensity, and (l) Raman spectra of detection system under different reaction time between target RNA (0.01 μM) and probe DNA. Parameter optimization experiments are completed at room temperature.
Appropriate DNA concentration is important to improve RNA detection sensitivity. To get appropriate DNA concentration, 85 μl AuNPs solution is incubated with different concentrations of DNA for 30 min and then 0.3 × SSC is introduced into the mixtures. The change in absorbance spectrum is presented in Fig. 3c. Then, the solution is centrifuged at 10,000 rpm at 4 °C for 15 min. After centrifugation, solid-liquid separation is performed, the solid phase is subjected to Raman detection, and the supernatant is subjected to fluorescence detection. As shown in Fig. 3e, when the DNA concentration decreases from 1 μM to 0.05 μM, Raman spectra show stronger representative peaks of Cy3 labeled on DNA probes. Fig. 3f illustrates that the fluorescence intensity of supernatant increases with the increase of DNA concentration higher than 0.05 μM, which indicates that there are excess DNA probes in the solution. When DNA concentration is lower than 0.05 μM, no fluorescence intensity is detected in the supernatant. Therefore, the appropriate concentration of DNA is 0.05 μM, which is enough to conjugate with AuNPs without obviously extra amount in the solution. The valid conjugation is supported by the red shift of the main peak at ∼520 nm and appearance of the secondary peak at ∼690 nm in the absorbance spectra, which is induced by the electromagnetic resonance on the surface of nanomaterials during the reaction with biomolecules [39,40].
DNA probes incubation time is another important parameter to optimize. 10 μl DNA probes of 0.05 μM are loaded into AuNPs solutions with different incubated time, and then 0.3 × SSC is introduced into the mixtures. As shown in Fig. 3g and h, the absorbance ratio of A690nm/A520nm decreases with the incubation time less than 30 min, and then remains substantially stable over time, indicating the best incubation time of 30 min to completely conjugate with AuNPs.
The influence of reaction time between DNA and RNA is also investigated. The absorbance measurements are carried out according to the procedure described above. Fig. 3i shows the absorbance spectrum dependence on the reaction time of P-ORF1 and ORF1, and Fig. 3j illustrates that the absorbance ratio of A690nm/A520nm gradually increases and reaches a steady state after 20 min. Then the reaction solution is centrifuged, and the supernatant is subjected to fluorescence detection. As shown in Fig. 3k, after the hybridization of DNA probe and target RNA for 20 min, the supernatant was separated form AuNPs through centrifugation for 15 min, and the fluorescence intensity in the supernatant was measured. Raman detection of the solid phase also confirmed the best DNA-RNA incubation time of 20 min as shown in Figure 3l. Therefore, incubation time of 20 min is applied to form RNA-DNA duplex.
3.3. Selective detection of target RNA with triple-mode biosensor
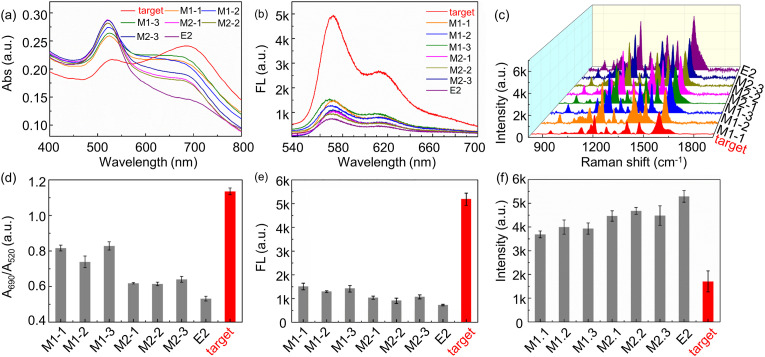
Selectivity is one of the most important parameters in RNA sensing. One and two base-pair-mismatched RNAs are selected to test the selectivity of the proposed biosensor, among which M1-1, M1-2, and M1-3 are RNAs with a single base mismatch at the 3′ end, 5’ end and the middle, compared with target RNA, respectively. M2-1, M2-2, and M2-3 are RNAs which have two-base mismatches from target gene at different positions, and E2 is RNA completely unrelated to target RNA. Under optimal conditions, the experimental results are shown in Fig. 4 . In the presence of target RNAs, RNA-DNA duplex forms, which leads to desorption of DNA probes form AuNPs and causes the aggregation of AuNPs. The experimental results indicates that RNAs with only one base mismatch is distinguished clearly by the proposed sensors. This strategy can discriminate single base differences among RNA targets. Absorbance, fluorescence, and Raman spectra of RNAs with only one-base mismatch has great difference from target RNA, as shown in Figure a-c, similarly to A690nm/A520nm ratio, fluorescence peak intensity, and representative Raman peak intensity in Figure d-f. RNAs with two-base mismatches have slight larger sensing signal difference from target RNAs than RNAs with one-base mismatches. The fluorescence intensity at 570 nm of target RNA has sensing signal 70% higher than non-specific RNA, and the representative Raman peak at 1195 cm−1 of target RNA is significally lower than other non-specific RNAs. Among the representative peaks of Cy3, the peak position at 1195 cm−1 is selected not only because of its obvious characteristics, but also because DNA probe has no characteristic peak at this place, which does not interfere with the experimental results [41] These results indicate that the selectivity of the proposed biosensor is excellent for COVID-19 RNA detection.
Fig. 4.
Selectivity investigation of proposed sensor under the optimized experimental conditions. (a) Absorption, (b) fluorescence, and (c) Raman spectra of sensing system in the presence of 10−8 M target RNA and other non-specific RNAs. (d) A690nm/A520nm Ratio, (e) fluorescence intensity at 570 nm, and (f) Raman intensity at 1195 cm−1 in the presence of different RNAs.
3.4. Detection sensitivity and stability of the triple-mode biosensor
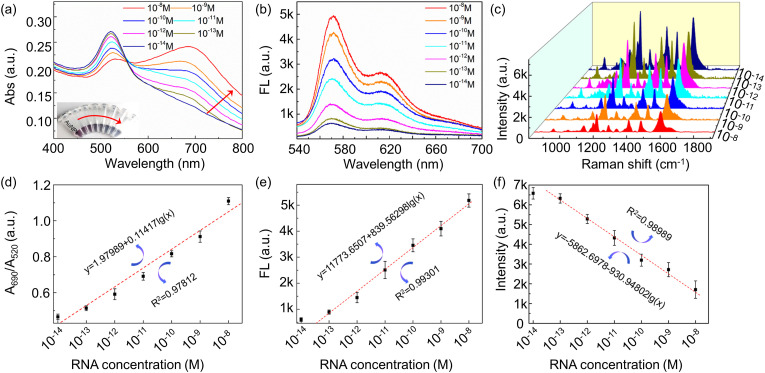
To evaluate the sensitivity of the triple-mode sensor, samples containing different concentrations of target RNA are detected under the optimized experimental conditions. As shown in Fig. 5 a, the characteristic absorption peak of AuNPs at 520 nm gradually decreases, the absorption peak at 690 nm gradually increases, and the color gradually becomes blue from red with the increase of RNA concentration. In addition, a red shift (570 → 620 nm) in representative fluorescence peak is present with the increase of RNA concentration, and the fluorescence peak intensity is greatly increased, as in Fig. 5b. While the Raman intensity decreases with increasing RNA concentration because of the consumption of DNA probe, as in Fig. 5c. More in detail, A690nm/A520nm ratio, fluorescence intensity at 570 nm, and Raman intensity at 1195 cm−1 present excellent linear relationship with target DNA concentration in Fig. 5d–f. The detection limit is calculated according to the formula LOD = 3S b1 /S [42,43], where Sb1 is the standard deviation of blank value, and S is the sensitivity at low concentration, that is, the slope of the standard working curve within the range of low concentration. After the calculation, the limit detection of absorbance is 0.58 pM, the limit detection of Raman is 2.17 pM, and the limit detection of fluorescence is 1.11 pM.
Fig. 5.
(a) Absorption, (b) fluorescence, and (c) Raman spectra of the triple-mode sensor in response to different concentrations of target RNA. Quantification relationship between target RNA concentration and (d) A690nm/A520nm ratio, (e) fluorescence intensity at 570 nm, (f) Raman intensity at 1195 cm−1.
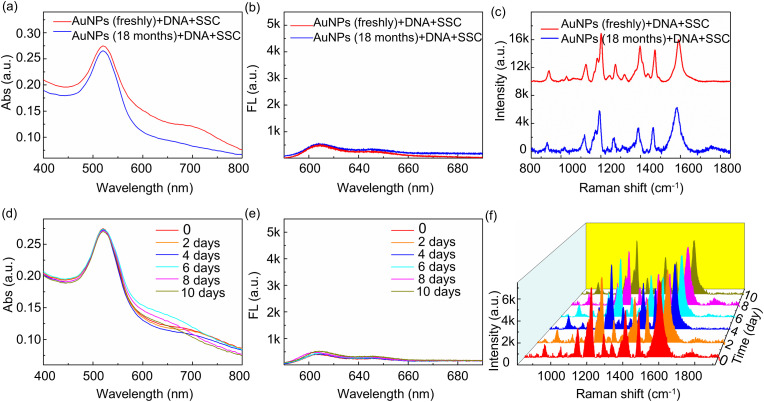
In order to test the stability of DNA probe conjugated AuNPs solutions, we added 10 μl 0.05 μM DNA probes into 85 μl AuNPs solution, and then added 5 μl 0.3 × SSC solution. The long-term stability is supported by the samples prepared 18 months ago, as shown in Fig. 6 a–c for all three modes, and there are negligible changes in the detection signals. At the same time, 10 similar samples were prepared to test the short-term stability of the DNA probe conjugated AuNPs solutions. The samples were subjected to three-signal analysis every day. The detection results are shown in Fig. 6d–f. The signals in three modes show good stability.
Fig. 6.
Absorbance (a), fluorescence (b) and Raman spectra (c) of AuNPs (freshly and 18 months ago) containing DNA probes upon the addition of 0.3 × SSC. After 0.3 × SSC was added, the absorbance (d), fluorescence (e) and Raman spectra (f) of AuNPs containing DNA probes were tested for stability within ten days.
3.5. Simulated samples test
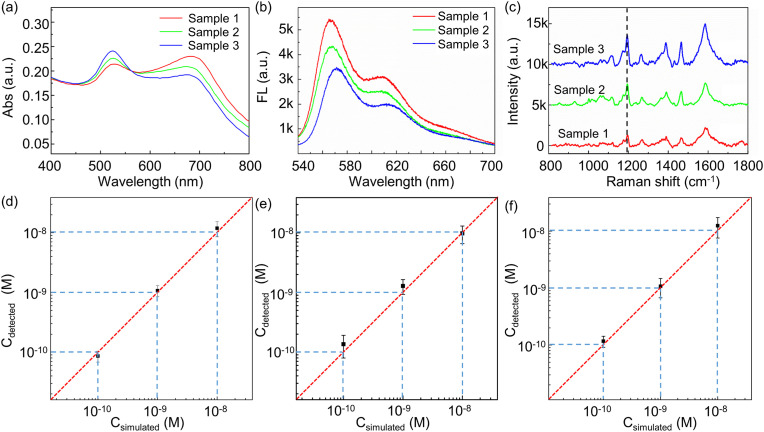
In order to simulate real sample detection, four target RNA segments with different concentrations are spiked in TE buffer as listed in Table 2 , and P-ORF1 probe is utilized in the reaction system. The concentration of target RNA varies from 10 nM to 0.1 nM, while the concentration of non-specific RNA- ORF1, E1, and E2 is kept at 1 nM. Fig. 7 a shows the absorbance spectrum of RNA detected in mixed samples. Fig. 7b and c show the fluorescence intensity and Raman intensity detected in mixed samples with different concentration of target RNA. With the increase in RNA concentration, the fluorescence intensity increased and the Raman intensity decreased. The detected concentrations are compared to spiked concentrations in Fig. 7d–f, and the detected concentrations are similar to the spiked concentrations, indicating that the triple-mode sensor is capable of detecting RNA with good recovery. The three detecting modes present the similar RNA level with deviation of 8.21%, which indicates the detection accuracy of the triple-mode sensor. And the calculated recovery efficiency of the sensor is 97–126%.
Table 2.
The concentration of 3 mixed RNA samples.
| RNA | Sample 1 | Sample 2 | Sample 3 |
|---|---|---|---|
| ORF1 | 10−8M | 10−9M | 10−10M |
| ORF2 | 10−9M | 10−9M | 10−9M |
| E1 | 10−9M | 10−9M | 10−9M |
| E2 | 10−9M | 10−9M | 10−9M |
Fig. 7.
Absorption (a), fluorescence (b), and Raman spectra (c) of the triple-mode sensor in response to different concentrations of RNA in mixed samples. (d)–(f) Comparison of detected and spiked RNA concentrations.
3.6. Multi-site probes improved detection performance of target RNA
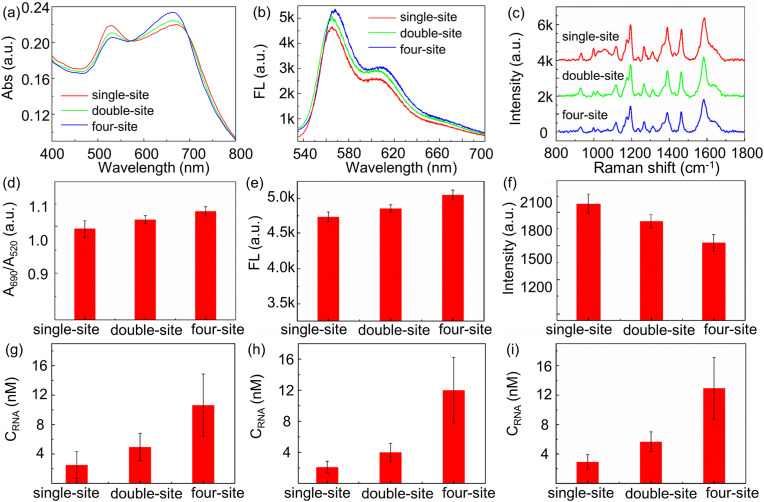
Single-point recognition of target RNA has serious risk of missed capture. As a result, four probes are utilized to recognize different specific sites in target RNAs to increase the accuracy of detection. The sample is prepared with ORF1ab and E segments mixture, and single-site probe P-ORF1, double-site probes P-ORF1and P-ORF2, four-site probes P-ORF1, P-ORF2, P-E1, and P-E2 are applied in the detection system. The concentration of each probe is 0.0125 μM in single-site, double-site and four-site probes system. Obviously, triple-mode signals enhance greatly with increased detection sites. Four-site probes detection system has much stronger detection signal than that of double-site and single-site probe detection, as shown in Fig. 8 . The absorbance A690nm/A520nm ratio and the fluorescence peak intensity increase, while the Raman peak reduces with increased detection sites, as shown in Fig. 8d–f. If the detection concentration of target RNA is calculated according to the relationship equation in Fig. 5d–f, the overall detected concentration from different detection sites are presented in Fig. 8g–i, which means that multiple sites probes detection enlarges detection signal by multiple times, because the probability to catch targets increases with detection sites.
Fig. 8.
(a) Absorption, (b) fluorescence, (c) Raman spectra in response to detection system with different site probes. (d) The absorbance A690nm/A520nm ratio, (e) fluorescence intensity at 570 nm, and (f) Raman intensity at 1195 cm−1 in response to detection system with different detection site probes. Overall detected concentration from (g) absorbance, (h) fluorescence, and (i) Raman spectra in detection system with different site probes.
3.7. The detection of simulated samples by triple-mode biosensor with four-site probes
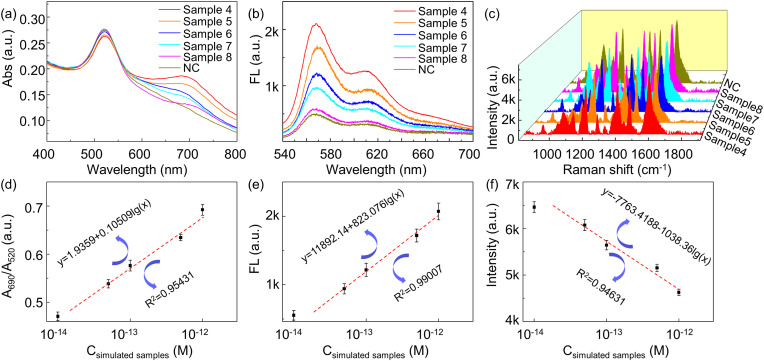
In the detection system with multiple-site probes, it is more sensitive because of higher hybridization opportunity at multiple detection sites. Four-site probes are utilized with a concentration of 0.0125 μM for each probe. In order to simulate real sample detection, four target RNA segments with different concentrations are spiked in TE buffer as listed in Table 3 , and their concentrations vary from 1 pM to 0.01 pM. The experimental results show that the detection sensitivity of the four-site probes system is higher than that of single-site probe system, and the detection signal is enhanced to 4 times as that of single-site system. Fig. 9 shows that the detection limit of the triple-mode biosensor with four-site probes is 160 fM in absorbance mode, 259 fM in fluorescence mode, and 395 fM in SERS mode. We believe that the sensitivity of the sensor platform could be improved further by designing more specific probes since the RNA length is more than 20 thousand bases.
Table 3.
The concentration of the mixed RNA samples.
| RNA | Sample 4 | Sample 5 | Sample 6 | Sample 7 | Sample 8 |
|---|---|---|---|---|---|
| ORF1 | 1pM | 0.5pM | 0.1pM | 0.05pM | 0.01pM |
| ORF2 | 1pM | 0.5pM | 0.1pM | 0.05pM | 0.01pM |
| E1 | 1pM | 0.5pM | 0.1pM | 0.05pM | 0.01pM |
| E2 | 1pM | 0.5pM | 0.1pM | 0.05pM | 0.01pM |
Fig. 9.
(a) Absorption, (b) fluorescence, and (c) Raman spectra of triple-mode biosensor in response to different concentrations of simulated samples. Quantification relationship between simulated samples concentration and (d) A690nm/A520nm ratio, (e) fluorescence intensity at 570 nm, (f) Raman intensity at 1195 cm−1. Note: NC is negative control sample.
The triple biosensor based on colorimetry, fluorescence, and SERS is capable of detecting SARS-CoV-2 viral RNA with high selectivity and low detection limit. The main parameters of different methods are listed in Table 4 .
Table 4.
Overview of SARS-CoV-2 detection methods.
| Method | Biomarker | Detection Limit | Detection Time | Amplification | References |
|---|---|---|---|---|---|
| Electrochemical immunosensor | Spike and nucleocapsid protein | 19 ng/mL and 8 ng/mL | 30min | no | [44] |
| FET biosensors | spike protein | 100 fg/ml | >1min | no | [8] |
| PCR | RdRp and N assays | 1.5 × 101 copy/ml | <3 h | yes | [45] |
| LAMP | ORF1a and N gene | 4.8 copy/μl | >30min | yes | [46] |
| CRISPR | ORF1ab and N regions | 2 copy/μl | ∼50min | yes | [47] |
| Dual-Functional Plasmonic Photothermal Biosensors | RdRp and E gene | 0.22pM | 800s | no | [48] |
| Triple-mode biosensor | RdRp and E gene | 160 fM | 40min | no | This work |
3.8. Advantages and limitations of the triple-mode biosensors
The developed triple-mode biosensor provides a few advantages for screening COVID-19 disease. The most important merit of the triple-mode biosensor is the detection accuracy through inter-proof results between three working modes, which reduces the false negative/positive reading of SARS-CoV-2 and avoids the spreading of the COVID-19. Another advantage of the triple-mode biosensor is its rapid process, which only needs 40 min for the whole detection process. The screening of large population is needed when the virus infection happens. Early and quick diagnosis of COVID-19 patient could save large amount of money through effective treatment and quarantining. Comparing to conventional PCR detection method, our triple-mode sensing system can save more time during the screening. Its process is simple because the long-term stability enables the well-prepared reaction solution ready for use any time. During practical application, it is optional to choose dual-mode biosensing -colorimetric and fluorescence-which are detected using the portable microplate reader and photoluminescence system. They conduct automatically scanning of all the samples in the microplates. The simple detection process and portable equipment are extremely important for population screening.
In addition, complicated RNA extraction process is not necessary during the proposed approach. During the reaction system, the concentration of salt plays important role. During practical detection, RNA extraction could be skipped if throat swab eluent with low salt is utilized. The viruses in the throat swab sample could be lysed through ultrasonication, which may break down the long RNA sequence randomly into short segments, but the short segments are suitable for our detection method even though partial targets may get broken. It is worth to note that the developed triple-mode biosensor can be used to detect any genes by using corresponding specific gene probes.
Despite the several distinct merits of the developed triple-mode biosensor, there are still some limitations associated with current sensing system. The major limitation is the sensitivity. The acceptable detection range (160 fM – 1 nM) of the current triple-mode biosensor is comparable to most developed existing gene biosensor, which may be suitable for the screening of COVID-19 patient with severe infection. But for patients with mild infection, the performance of our triple-mode biosensor needs to be improved by incorporating gene amplification processing. Recently, Yan [49] and Xing [50] reported the signal amplification strategy by using fluorescence molecular amplification technology, resulting in the ultrasensitive gene detection at the level of ∼aM. We believe that the proposed triple-mode biosensor could reach much lower detection limit by incorporating the fluorescence signal amplification strategy.
4. Conclusion
In summary, we fabricated a facile genes sensing platform based on AuNPs for triple-mode RNA biosensing, which is achieved by integrating colorimetric, SERS and fluorescence detection. Due to the unique optical properties and good Raman enhancement properties of AuNPs, visible colorimetric sensing and sensitive SERS detection could be carried out. Meanwhile, due to the different adsorption behaviors of AuNPs to single and double chains, fluorescence signal response is obtained from the supernatant after centrifugation, and the triple mode detection of RNA in SARS-CoV-2 is successfully realized. Through the coupling of multiple-site probes in the sensor system, detection accuracy and sensitivity of the tripe-mode biosensor are improved, and the false negative of the detected signal could be greatly reduced. The proposed work shows potential applications in the screening of COVID-19 infection with reduced false negative, as well as accurate early diagnosis and real-time monitoring of patients infected with viruses.
CRediT authorship contribution statement
Yakun Gao: carried out the experiments and wrote this manuscript. Yingkuan Han: conducted AuNPs characterization. Chao Wang: helped perform the experiments. Le Qiang: helped perform the experiments. Jianwei Gao: helped to do Raman tests. Yanhao Wang: helped to do Raman tests. Hong Liu: helped revise the manuscript. Lin Han: designed the study. Yu Zhang: brought up the idea.
Declaration of competing interest
All the authors declare no conflicts of interest with this work.
Acknowledgement
This work was supported by the National Key R&D Plan of China (Grant No. 2017YFB0405400), the Major Innovation Projects in Shandong Province (2018YFJH0503), Natural Science Foundation for Distinguished Young Scientist of Shandong Province (Grant No. JQ201814), Natural Science Foundation for Young Scientists of China (Grant No.61701282), Qilu Young Scholar Funds (11500086963060), Shandong University multidisciplinary research and innovation team of young scholars, and Collaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong. The authors thank Xiaoju Li from State Key Laboratory of Microbial Technology, Shandong University for the assistance in transmission electron microscope (TEM) observation.
References
- 1.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. J. Am. Med. Assoc. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. 10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogoch, Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J. Trav. Med. 2020;27(2):1–3. doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W., Du R., Li B., Zheng X., Yang X., Hu B., Wang Y., Xiao G., Yan B., Shi Z. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 9.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 10.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brunink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corman V.M., Eckerle I., Bleicker T., Zaki A.M., Landt O., Eschbachbludau M., S V.B., Gopal R., Ballhause M., Bestebroer T.M. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39) doi: 10.2807/ese.17.39.20285-en. 20285. [DOI] [PubMed] [Google Scholar]
- 12.Thi V.L.D., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12(556):1–13. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microb. Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu M., Wang D., Wang H., Zhang X., Liang T., Dai J., Li M., Zhang J., Zhang K., Xu D. COVID-19 diagnostic testing: technology perspective. Clin. Transl. Med. 2020;10(4):e158. doi: 10.1002/ctm2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam J., Jung I.-B., Kim B., Lee S.-M., Kim S.-E., Lee K.-N., Shin D.-S. A colorimetric hydrogel biosensor for rapid detection of nitrite ions. Sensor. Actuator. B Chem. 2018;270:112–118. [Google Scholar]
- 16.Tian J., Zhang Y., Zhu L., Tian H., Li K., Huang K., Luo Y., Zhang X., Xu W. dsDNA/ssDNA-switchable isothermal colorimetric biosensor based on a universal primer and λ exonuclease. Sensor. Actuator. B Chem. 2020;323 128674. [Google Scholar]
- 17.Wang Z.Q., Wu S.S., Ciacchi L.C., Wei G. Graphene-based nanoplatforms for surface-enhanced Raman scattering sensing. Analyst. 2018;143(21):5074–5089. doi: 10.1039/c8an01266k. [DOI] [PubMed] [Google Scholar]
- 18.Chen R., Liu B., Ni H., Chang N., Luan C., Ge Q., Dong J., Zhao X. Vertical flow assays based on core–shell SERS nanotags for multiplex prostate cancer biomarker detection. Analyst. 2019;144(13):4051–4059. doi: 10.1039/c9an00733d. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Wu T., Yang Y., Wen Y., Wang S., Xu L.-P. Superwettable electrochemical biosensor based on a dual-DNA walker strategy for sensitive E. coli O157: H7 DNA detection. Sensor. Actuator. B Chem. 2020;321 128472. [Google Scholar]
- 20.Wang Y.-H., He L.-L., Huang K.-J., Chen Y.-X., Wang S.-Y., Liu Z.-H., Li D. Recent advances in nanomaterial-based electrochemical and optical sensing platforms for microRNA assays. Analyst. 2019;144(9):2849–2866. doi: 10.1039/c9an00081j. [DOI] [PubMed] [Google Scholar]
- 21.Špringer T., Hemmerová E., Finocchiaro G., Krištofiková Z., Vyhnálek M., Homola J. Surface plasmon resonance biosensor for the detection of tau-amyloid β complex. Sensor. Actuator. B Chem. 2020;316 128146. [Google Scholar]
- 22.Castiello F.R., Tabrizian M. Gold nanoparticle amplification strategies for multiplex SPRi-based immunosensing of human pancreatic islet hormones. Analyst. 2019;144(8):2541–2549. doi: 10.1039/c9an00140a. [DOI] [PubMed] [Google Scholar]
- 23.Yang Q., Wang X., Peng H., Arabi M., Li J., Xiong H., Choo J., Chen L. Ratiometric fluorescence and colorimetry dual-mode assay based on manganese dioxide nanosheets for visual detection of alkaline phosphatase activity. Sensor. Actuator. B Chem. 2020;302 127176. [Google Scholar]
- 24.Niu T., Yu T., Yin G., Chen H., Yin P., Li H. A novel colorimetric and ratiometric fluorescent probe for sensing SO 2 derivatives and their bio-imaging in living cells. Analyst. 2019;144(5):1546–1554. doi: 10.1039/c8an02331j. [DOI] [PubMed] [Google Scholar]
- 25.Masterson A.N., Liyanage T., Berman C., Kaimakliotis H., Johnson M., Sardar R. A novel liquid biopsy-based approach for highly specific cancer diagnostics: mitigating false responses in assaying patient plasma-derived circulating microRNAs through combined SERS and plasmon-enhanced fluorescence analyses. Analyst. 2020;145(12):4173–4180. doi: 10.1039/d0an00538j. [DOI] [PubMed] [Google Scholar]
- 26.Qu L.-L., He S.-H., Wang J.-J., Lin Z.-C., Barry D., Yang G.-H., Wang P., Zhang P., Li H.-T. Fluorescence-surface enhanced Raman scattering dual-mode nanosensors to monitor hydroxyl radicals in living cells. Sensor. Actuator. B Chem. 2017;251:934–941. [Google Scholar]
- 27.Li D., Li C., Liang A., Jiang Z. SERS and fluorescence dual-mode sensing trace hemin and K+ based on G-quarplex/hemin DNAzyme catalytic amplification. Sensor. Actuator. B Chem. 2019;297 126799. [Google Scholar]
- 28.Song C., Li J., Sun Y., Jiang X., Zhang J., Dong C., Wang L. Colorimetric/SERS dual-mode detection of mercury ion via SERS-Active peroxidase-like Au@AgPt NPs. Sensor. Actuator. B Chem. 2020;310 127846. [Google Scholar]
- 29.Shang H., Xu H., Jin L., Chen C., Song T., Wang C., Du Y. Electrochemical-photoelectrochemical dual-mode sensing platform based on advanced Cu9S8/polypyrrole/ZIF-67 heterojunction nanohybrid for the robust and selective detection of hydrogen sulfide. Sensor. Actuator. B Chem. 2019;301 127060. [Google Scholar]
- 30.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyrlaki I., Ekman M., Lentini A., Rufino de Sousa N., Papanicolaou N., Vondracek M., Aarum J., Safari H., Muradrasoli S., Rothfuchs A.G., Albert J., Hogberg B., Reinius B. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 2020;11(1):4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storhoff J.J., Elghanian R., Mucic R.C., Mirkin C.A., Letsinger R.L. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998;120(9):1959–1964. [Google Scholar]
- 33.Liu J. Adsorption of DNA onto gold nanoparticles and graphene oxide: surface science and applications. Phys. Chem. Chem. Phys. 2012;14(30):10485–10496. doi: 10.1039/c2cp41186e. [DOI] [PubMed] [Google Scholar]
- 34.Miao P., Tang Y., Mao Z., Liu Y. Adamantane derivatives functionalized gold nanoparticles for colorimetric detection of MiRNA. Part. Part. Syst. Char. 2017;34(6) 16400405. [Google Scholar]
- 35.Stobiecka M., Deeb J., Hepel M. Ligand exchange effects in gold nanoparticle assembly induced by oxidative stress biomarkers: homocysteine and cysteine. Biophys. Chem. 2010;146(2–3):98–107. doi: 10.1016/j.bpc.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Stobiecka M., Hepel M. Double-shell gold nanoparticle-based DNA-carriers with poly-L-lysine binding surface. Biomaterials. 2011;32(12):3312–3321. doi: 10.1016/j.biomaterials.2010.12.064. [DOI] [PubMed] [Google Scholar]
- 37.Sun M., Liu F., Zhu Y., Wang W., Hu J., Liu J., Dai Z., Wang K., Wei Y., Bai J. Salt-induced aggregation of gold nanoparticles for photoacoustic imaging and photothermal therapy of cancer. Nanoscale. 2016;8(8):4452–4457. doi: 10.1039/c6nr00056h. [DOI] [PubMed] [Google Scholar]
- 38.Pamies R., Cifre J.G.H., Espín V.F., Collado-González M., Baños F.G.D. J.G. de la Torre, Aggregation behaviour of gold nanoparticles in saline aqueous media. J. Nanoparticle Res. 2014;16(4):2376. [Google Scholar]
- 39.Philip D. Synthesis and spectroscopic characterization of gold nanoparticles. Spectrochim. Acta Mol. Biomol. Spectrosc. 2008;71(1):80–85. doi: 10.1016/j.saa.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Shokri E., Hosseini M., Davari M.D., Ganjali M.R., Peppelenbosch M.P., Rezaee F. Disulfide-induced self-assembled targets: a novel strategy for the label free colorimetric detection of DNAs/RNAs via unmodified gold nanoparticles. Sci. Rep. 2017;7(1) doi: 10.1038/srep45837. 45837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barhoumi A., Zhang D., Tam F., Halas N.J. Surface-enhanced Raman spectroscopy of DNA. J. Am. Chem. Soc. 2008;130(16):5523–5529. doi: 10.1021/ja800023j. [DOI] [PubMed] [Google Scholar]
- 42.Kim W.H., Lee J.U., Song S., Kim S., Choi Y.J., Sim S.J. A label-free, ultra-highly sensitive and multiplexed SERS nanoplasmonic biosensor for miRNA detection using a head-flocked gold nanopillar. Analyst. 2019;144(5):1768–1776. doi: 10.1039/c8an01745j. [DOI] [PubMed] [Google Scholar]
- 43.Xia Y., Wang L., Li J., Chen X., Lan J., Yan A., Lei Y., Yang S., Yang H., Chen J. A ratiometric fluorescent bioprobe based on carbon dots and acridone derivate for signal amplification detection exosomal microRNA. Anal. Chem. 2018;90(15):8969–8976. doi: 10.1021/acs.analchem.8b01143. [DOI] [PubMed] [Google Scholar]
- 44.Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D’Amore N., Regalbuto E., Salvatori P. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2020;171 doi: 10.1016/j.bios.2020.112686. 112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim S., Park E.C., Park D., Lee J.-H., Byeon C.W. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. BioRxiv. 2020;6:2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. medRxiv; 2020. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. 2020.02.26.20028373. [Google Scholar]
- 47.Huang Z., Tian D., Liu Y., Lin Z., Lyon C., Lai W., Fusco D., Drouin A., Yin X., Hu T. Ultra-sensitive and high-throughput CRISPR-Powered COVID-19 diagnosis. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112316. 112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 49.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing W., Liu Y., Wang H., Li S., Lin Y., Chen L., Zhao Y., Chao S., Huang X., Ge S., Deng T., Zhao T., Li B., Wang H., Wang L., Song Y., Jin R., He J., Zhao X., Liu P., Li W., Cheng J. A high-throughput, multi-index isothermal amplification platform for rapid detection of 19 types of common respiratory viruses including SARS-CoV-2. Engineering. 2020;6:1130–1140. doi: 10.1016/j.eng.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]