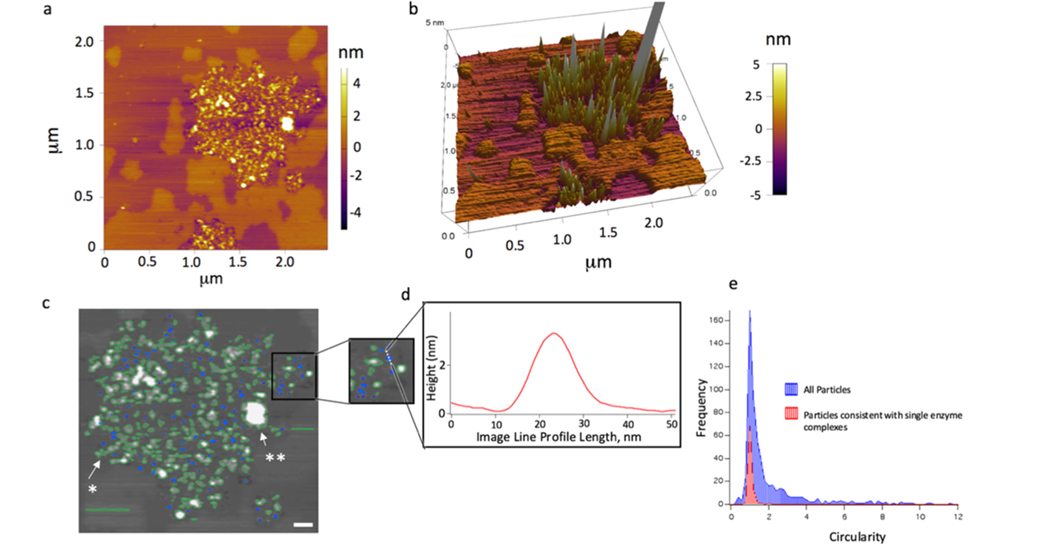
Figure 3.
Feature volume analysis of AFM height images of the γ-secretase complex. (a) Dispersion of enzyme complexes within a larger area at longer incubation time (t = 283 min of incubation time at 25 °C). (b) 3D rendering graph of dispersed single enzyme complexes and their positioning in both the Lo and Ld phases. (c) Overlaid images with the results of a particle localization analysis tool in Igor Pro (blue and green overlays). (d) Inset is zoomed in to a smaller ROI and an image line profile of the dotted line in the inset. The feature shown has an approximate volume of 149.5 nm3. The blue labeled features are small, circular, and consistent with single γ-secretase complexes (scale bar 100 nm). To choose the features consistent with the single enzyme complex, the circularity was chosen as a filter (circularity: the ratio of the square of the perimeter to (4 × π × area)). This value approaches 1 for a perfect circle. (e) Histogram of the single enzyme feature circularity (red) with the histogram of all of the features present in a 4 × 4 μm2 image (blue).