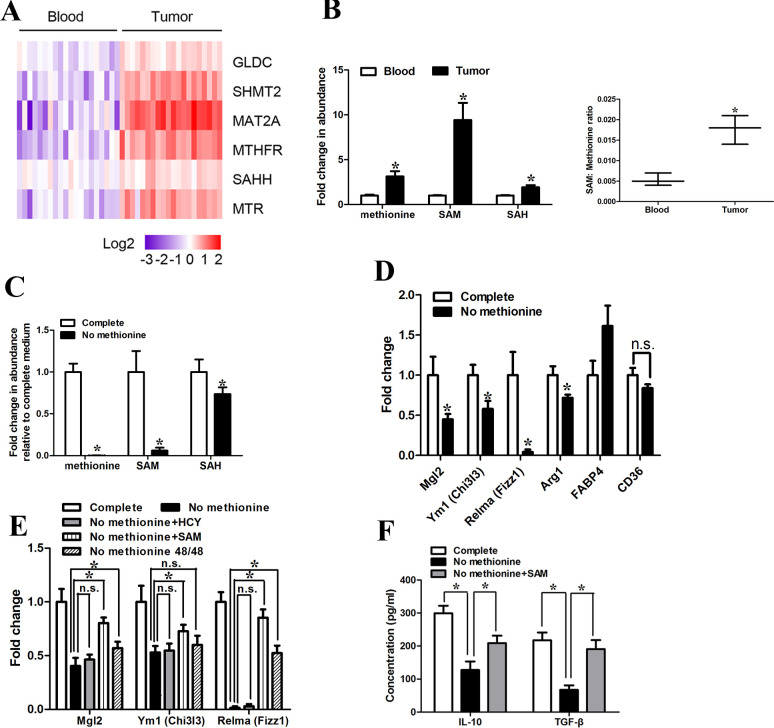
Figure 1.
Tumor-infiltrating macrophages of GC had enhanced levels of methionine metabolism activity. (A) CD14+ cells were purified from the tumor tissues and peripheral blood of five patients with GC. The expression levels of methionine metabolism-related gene were quantified by qRT-PCR. (B) (Left) LC-MS was used to determine abundance of intracellular primary methionine cycle metabolites. Values were normalized to that in CD14+ cells purified from peripheral blood. (Right) Ratio of SAM to methionine levels in CD14+ cells purified from peripheral blood and tumor tissues. (C) LC-MS was used to determine abundance of methionine cycle metabolites in CD14+ cells purified from tumor tissues 48 hours after methionine starvation. Values were normalized to that in the complete condition. (D) qPCR analysis of the M2-associated genes Relma, Mgl2, Ym1, Fabp4, Arg1 and CD36 48 hours after methionine starvation. (E) CD14+ cells purified from tumor tissues were starved for 48 hours for methionine but supplemented with homocysteine (HCY; 250 µM), SAM (500 µM) or replated into complete medium for the next 48 hours (48/48). qPCR analysis of the M2-associated genes Relma, Mgl2, Ym1. (F) CD14+ cells purified from tumor tissues were starved for 48 hours for methionine but supplemented with SAM (500 µM). The levels of IL-10 and TGF-β production were determined by ELISA. Data are presented as the mean±SD; *p<0.05. N=3 biologically independent experiments. GC, gastric cancer; SAM, S-adenosylmethionine.