Fig. 1. DLSμR overview.
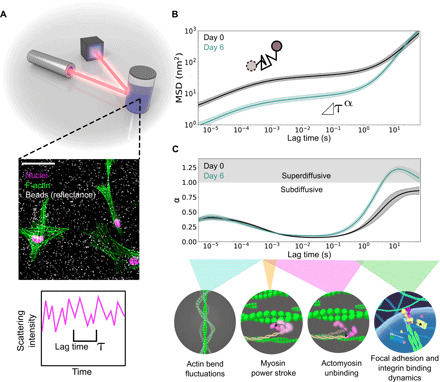
(A) Schematic of DLSμR. Mammalian cells are encapsulated within a 3D extracellular matrix containing polystyrene tracer particles. Tracer particle dynamics are extracted by measuring light scattering fluctuations across different time scales, τ. The middle inset is a confocal microscopy image of HMFs 6 days after encapsulation in a mixture of col/rBM. F-actin fibers (green false color) were stained with tetramethyl rhodamine B isothiocyanate (TRITC)–phalloidin. Nuclei (magenta false color) were counterstained with DAPI (4′,6-diamidino-2-phenylindole). Polystyrene beads (false-colored white) were visualized by confocal reflectance from a 635-nm laser. Scale bar, 50 μm. (B) Tracer particle MSD as a function of lag time obtained from DLSμR measurements with HMFs in col/rBM. Days 0 and 6 indicate the time elapsed since encapsulation. (C) Local power-law scaling exponent of particle motion, α, as a function of lag time. The bottom schematic illustrates processes corresponding to different time scale regimes: F-actin bend fluctuations (4, 5), the myosin power stroke (6), myosin unbinding lifetimes (47, 48), focal adhesion turnover (7), and integrin binding lifetimes (49). In (B) and (C), curves represent the geometric and arithmetic means among biological replicates (n = 7), respectively. Shaded bands represent 68% confidence intervals of the respective means.
