Fig. 3. Establishment of the R loop CUT&Tag for native R loop mapping.
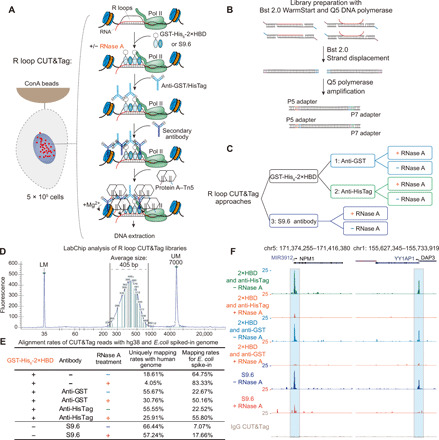
(A) Overview of the R loop CUT&Tag workflow. Cells were immobilized on concanavalin A (ConA)–coated magnetic beads, followed by cell permeabilization. GST-His6-2×HBD or S9.6 is used to recognize the R loops in the presence or absence of RNase A. Anti-GST, anti-HisTag, or secondary antibodies were applied to enhance the tethering of pA-Tn5 transposome at the GST-His6-2×HBD or S9.6-bound sites. After extensive wash, the pA-Tn5 transposome is activated to integrate the adapters on the chromatin. (B) CUT&Tag library preparation with Bst 2.0 WarmStart and Q5 polymerase. Strand displacement was performed with Bst 2.0, followed by library amplification with Q5 DNA polymerase. (C) Three different approaches for R loop CUT&Tag analysis. (D) LabChip analysis of R loop CUT&Tag library demonstrating the library size ranges from 220 to 700 bp with an average size of 405 bp. UM, upper marker; LM, lower marker. (E) Alignment rates of R loop CUT&Tag reads to the human hg38 and E. coli spiked-in genomes. RNase A treatment markedly decreases the alignment rates of CUT&Tag reads to the human genome, suggesting the specificity of GST-His6-2×HBD and S9.6 on R loop recognition. (F) UCSC genome browser tracks of CUT&Tag signals at the NPM1 and YY1AP1 loci. The tracks were normalized by reads per million, and the RNase A–treated groups were further normalized with the E. coli spike-in control.
