Fig. 5. R loop CUT&Tag signals are sensitive to RNase H digestion.
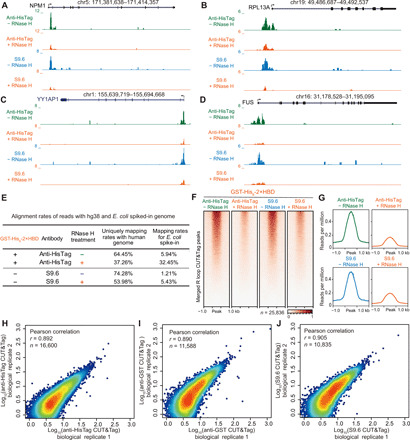
(A to D) UCSC genome browser tracks of CUT&Tag signals at the NPM1, RPL13A, YY1AP1, and FUS loci. The tracks were normalized by reads per million and the RNase H–treated groups were further normalized with the E. coli spike-in control. (E) Alignment rates of R loop CUT&Tag reads to the human hg38 and E. coli spiked-in genomes. Four-hour RNase H treatment markedly reduces the alignment rates of CUT&Tag reads to the human genome and increases the alignment rates of reads to E. coli spiked-in genomes. (F and G) Heatmap and metaplot analysis of R loop CUT&Tag signals at all of the peaks from GST-His6-2×HBD and S9.6 CUT&Tag. RNase H digestion markedly decreases the CUT&Tag signals at those peaks, demonstrating great specificity of GST-His6-2×HBD and S9.6 on R loop recognition. (H to J) Reproducibility of R loop CUT&Tag methods. Biological replicates were performed, and the Pearson correlation was calculated with the reads per million at R loop peaks.
