Fig. 8. R loop signals are affected by ex vivo and in situ detecting strategies.
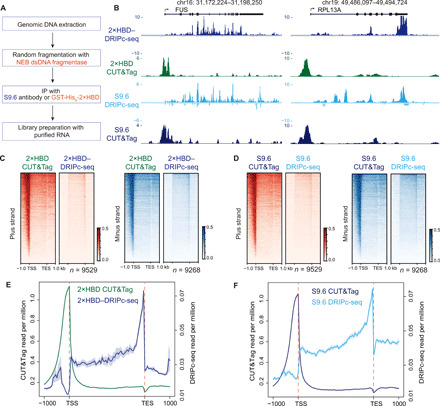
(A) Workflow of DRIPc-seq with GST-His6-2×HBD or S9.6 combined with random fragmentation of genomic DNA by NEB dsDNA fragmentase. IP, immunoprecipitation. (B) Genome browser tracks of DRIPc-seq and R loop CUT&Tag coverage at the FUS and RPL13A loci detected by GST-His6-2×HBD and S9.6. Signals were normalized by reads per million. (C and D) Heatmap analysis of DRIPc-seq (ex vivo) and R loop CUT&Tag (native, in situ) at all the protein-coding genes by GST-His6-2×HBD (C) or S9.6 (D). (E and F) Metagene plots of DRIPc-seq and R loop CUT&Tag by GST-His6-2×HBD (E) or S9.6 (F).
