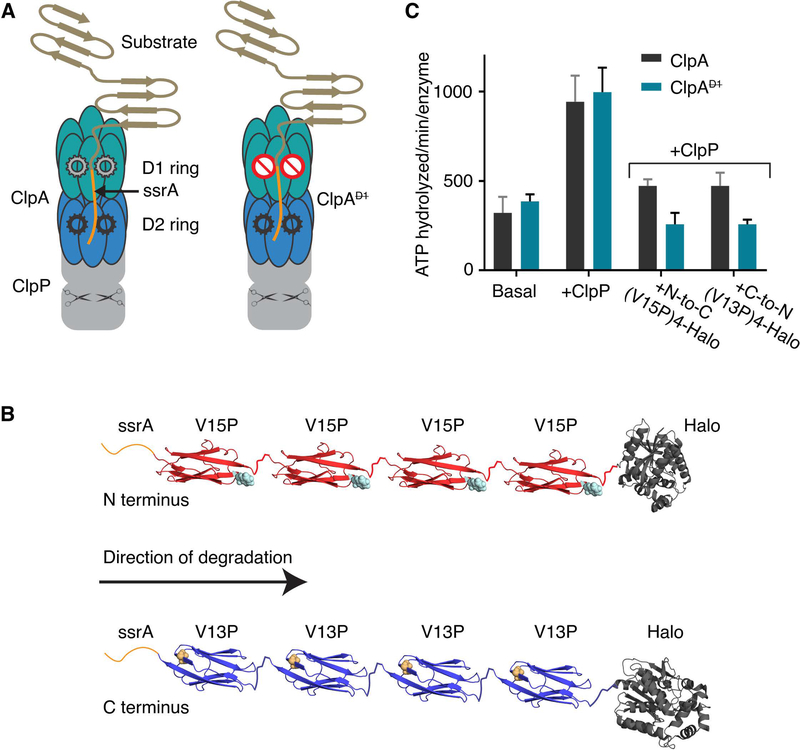
Figure 1: ClpAP protease, multidomain substrates, and ATP hydrolysis.
(A) Cartoon of ClpAP and ClpAP. Each ClpA subunit contains two AAA+ ATPase modules that form the D1 (teal) and the D2 (blue) rings in the assembled hexamer. ClpP (gray) is a barrel-shaped serine peptidase; scissors schematically represent peptidase active sites. A model protein substrate engaged for unfolding in the ClpA pore is shown. ClpA (right) contains a ⦰ symbol in the D1 ring to indicate an ATP-hydrolysis defect. (B) Schematic of ssrA-tagged multidomain substrates. The ssrA tag (orange) is present at the N-terminus of ssrA-(V15P)4-Halo (red, top) or the C-terminus of Halo-(V13P)4-ssrA (blue). The sites of the V15P and V13P mutations in the titin I27 domain are represented as spheres (note the titin domains are in opposite orientations). The Halo domain, used for covalent linkage to DNA for optical trapping, is dark gray. (C) Steady-state rates of ATP hydrolysis. Rates for ClpA or ClpA (0.1 μM hexamer) were measured in the absence of ClpP and substrate (basal), in the presence of ClpP (0.2 μM tetradecamer), or in presence of ClpP (0.2 μM tetradecamer) and substrate (5 μM). Values are averages ± 1 SD of three independent measurements.