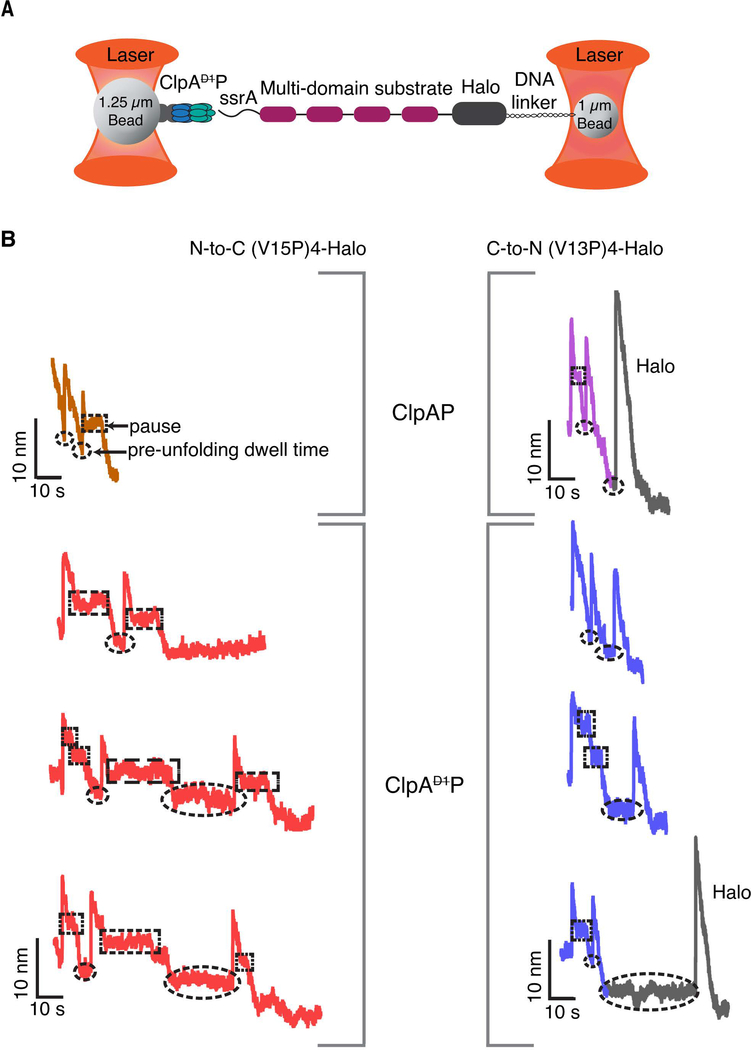
Figure 2: Single-molecule degradation by ClpAP and ClpAP.
(A) Optical-trap setup with ClpA attached to one bead via ClpPplatform and a multi-domain substrate attached to the smaller bead via linkage of the Halo domain to DNA. (B) Representative single-molecule degradation traces for ClpAP and ClpAP in the N-to-C direction (left) or C-to-N direction (right). The Halo domains are shown in gray. Rectangular and elliptical boxes mark pauses and pre-unfolding dwell times, respectively. Traces were recorded at forces between 8 and18 pN and were decimated to 300 Hz. ClpAP traces are from Olivares et al (Olivares et al., 2017; Olivares et al., 2014) and are shown for comparison.